Abstract
Titin (also known as connectin) is a giant protein with a wide range of cellular functions, including providing muscle cells with elasticity. Its physiological extension is largely derived from the PEVK segment, rich in proline (P), glutamate (E), valine (V), and lysine (K) residues. We studied recombinant PEVK molecules containing the two conserved elements: ≈28-residue PEVK repeats and E-rich motifs. Single molecule experiments revealed that calcium-induced conformational changes reduce the bending rigidity of the PEVK fragments, and site-directed mutagenesis identified four glutamate residues in the E-rich motif that was studied (exon 129), as critical for this process. Experiments with muscle fibers showed that titin-based tension is calcium responsive. We propose that the PEVK segment contains E-rich motifs that render titin a calcium-dependent molecular spring that adapts to the physiological state of the cell.
Titin comprises a multifunctional myofilament system in muscle, with a single molecule spanning the half-sarcomere (1-5). A large portion of the molecule functions as a molecular spring that, when extended, develops force. This force underlies the passive muscle force, which maintains the structural integrity of the contracting sarcomere and influences the filling behavior of the heart (3). Physiological force levels are largely determined by extension of the proline-glutamate-valine-lysine (PEVK) rich segment (6, 7). PEVK-like sequences are found in many titin-like proteins among evolutionary divergent organisms (8-11). The PEVK region of the human titin gene contains 114 exons; most code for conserved ≈28-residue PEVK repeats and 10 are more complex and encode E-rich motifs (12, 13). Here, we studied recombinant proteins that contain both PEVK repeats and glutamate (E)-rich motifs. Previous work suggests that the PEVK segment binds calcium with high affinity, raising the possibility that the extensibility of the PEVK segment may be calcium regulated (14). Thus, an important goal was to examine the effect of calcium on the mechanical behavior of the PEVK segment.
Methods
Proteins. We engineered only PEVK sequences (“naked” PEVK fragment) or PEVK sequences flanked by Ig-like domains (PEVK-Ig fusion fragment). Fragments were cloned, expressed, and purified by using routine methods (12, 15). All fragments were His-tagged at their amino terminus and, except when intended for structural analysis, had two Cys residues at the carboxyl terminus for covalent attachment to gold-coated substrates. Purified proteins were dialyzed into AB buffer (in mM; 25 Mops, pH 7.4/150 KCl/1 EGTA/1 DTT), quick frozen, and stored at -80°C. For additional details, see Supporting Text, which is published as supporting information on the PNAS web site.
Single Molecule Mechanics. Molecules were stretched by using an atomic force microscope specialized for stretching molecules (15, 16). Proteins (≈100 μg/ml) were allowed to bind for ≈10 min to gold-coated microscope slides (precleaned glass slides and freshly cleaved mica were also used; see text). Unbound molecules were washed away with buffer (in mM; 25 imidazole/200 KCl/4 MgCl2/1 EGTA, pH 7.4) to which CaCl2 was added to obtain free [Ca2+] ranging from ≈0.001 to 1,000 μM. Molecules were stretched by pressing a gold-coated cantilever (MSCT-AUHW: sharpened silicon nitride gold-coated cantilevers; Veeco Metrology Group, Santa Barbara, CA) against a PEVK-coated surface, pulling the cantilever away with a constant rate (500 nm/s) and, when reaching a predetermined amplitude, moving the cantilever back toward the surface. Force (F) was obtained from cantilever bending (Δd)as F = KΔd where K is cantilever stiffness. Stiffness was obtained for each unloaded cantilever by measuring its mean thermally driven vertical bending (d0) and applying the equipartition theorem: K 〈Δd02〉 = kBT (kB = Boltzmann's constant; T = absolute temperature). Cantilever stiffness was ≈10 pN/nm, and rms force noise (1-kHz bandwidth) was ≈5 pN. The zero-length, zero-force data point was obtained from the force response that corresponded to the cantilever tip reaching (or departing from) the substrate surface. The end-to-end length (z) of the tethered molecule was calculated by correcting the cantilever base displacement (s) with cantilever bending as z = s - F/K.
For naked PEVK fragments, analysis was restricted to molecules that, when stretched to ≈90% of theoretical contour length, remained attached and, thus, that gave a release curve. Due to irregularities in the initial part of the stretch curve (see Results and Discussion), the contour length (CL) and persistence length (PL) were determined by fitting the release data with the worm-like chain (WLC) equation (17). PLs were plotted in histograms, and, by using cluster analysis (fastclus, SAS Institute, Cary, NC), peak positions were obtained. For each experimental condition, we typically studied six preparations, determined for each the histogram peak positions (see Results and Discussion), and calculated the mean and SE of the obtained values. The experimentally determined CL was typically slightly less than the theoretical CL. For example, for fragment WT 119-136, the measured CL was 165 ± 13 nm whereas the theoretical CL is 184 nm (485 residues times 0.38 nm). Thus, ≈50 residues at the ends (terminal PEVK repeats) were not sampled. As for PEVK-Ig fusion proteins, these were also stretched with a velocity of 500 nm/s, and the unfolding characteristics of the Ig domains during stretch were used as a single molecule “fingerprint,” ensuring that single molecules were analyzed and that the full PEVK segment was stretched (see Results and Discussion). After fingerprinting, the release curve is dominated by unfolded Ig domains, and, thus, the release was not suitable for determining PEVK's PL. We performed “double stretch-release protocols” with a first stretch-release with limited amplitude (to avoid Ig unfolding) and providing a release curve for analysis, immediately followed by a large amplitude stretch to obtain the single molecule fingerprint. However, these experiments had a low success rate (force at the end of the initial stretch was either too low to allow analysis of the subsequent release curve, or force was too high and caused unfolding). Hence, we focused on the large amplitude stretch that resulted in unfolding of all domains and determined the PL of the force curve before the first unfolding peak. The mean and SE of all PL values obtained from recordings with the single molecule fingerprint were calculated.
Fluorescence. Emission spectra were recorded at 25°C with an ATF105 spectrofluorometer (Aviv Associates, Lakewood, NJ; λex = 290 nm; λem = 305-400 nm) by using 10 μM recombinant protein in AB buffer before and after adding small volumes of MgCl2 or CaCl2 stock solutions. (Free ion composition was calculated as in ref. 18.) Changes in the fluorescence intensity at λmax were determined and plotted against free [Ca2+], expressed as pCa (-log[Ca2+]); results were fit with a Hill equation to determine the [Ca2+] at which the change was half maximal.
Ultrasonic Velocimetry. As an alternative to fluorescence, we measured for solutions that contained PEVK fragments the ultrasonic velocity, a characteristic that can be used to assess changes in protein hydration and conformation (19) without requiring the presence of intrinsic fluorescent fluorophores. (This allowed PEVK repeats to be studied; see Results and Discussion.) A ResoScan System RU21 (Resonic Instruments AG, Ditzingen, Germany) was used with an ultrasonic resonator system [8 MHz, with two independent resonator cells (volume 180 μl)] that measures the absolute ultrasonic velocity in both cells with a resolution better than 10-6. An equal volume of CaCl2 stock was added to both the cell with protein (3 μM in AB buffer) and the reference cell (AB buffer). The velocity difference between the cells was determined (25.0°C), accounting for dilution due to CaCl2 addition.
Muscle Fiber Mechanics. Soleus fibers were dissected from mice, following approved methods [Institutional Animal Care and Use Committees (IACUC) 3095]. Skinned fibers (length ≈ 1.5 mm) were incubated in a relaxing solution-filled chamber and mounted to a force transducer and a motor; sarcomere length (SL) was measured with laser diffraction (for details see refs. 18, 20, and 21). Fibers were incubated with gelsolin fragment FX-45 (≈0.5 mg/ml) at 4°C for ≈50 h during continuous agitation (22). SDS/PAGE revealed that this treatment removed ≈95% of actin, and immunofluorescence revealed that the remaining actin was located in the Z-disk region of the sarcomere. Fibers were activated with pCa 4 solution at SL 2.1 μm to measure maximal active tension. Control fibers developed 123 ± 22 mN/mm2, and gelsolin-extracted fibers developed (at SL 2.1 μm) no detectable tension. The force-SL relation was then measured in pCa 9 solution and pCa 4 solution, both in the presence and absence of 30 mM 2,3-butanedione monoxime (BDM) to inhibit active tension (23). For stretches that took place at pCa 4, fibers were preincubated with calcium for several minutes. Fibers were stretched with a constant velocity that was either 0.1 or 10 μm per half sarcomere sec, after which the fibers were released back to the slack length. After a 20-min rest (pCa 9), a new stretch protocol was imposed. Provided maximal SL did not exceed ≈3.1 μm for the slow stretch, and ≈2.7 μm for the fast stretch, reproducible results were obtained. (Reaching longer SLs resulted in rundown. Whether this is due to an adverse effect of long-term gelsolin treatment on titin remains to be established.) We determined the tension-SL relation during the slow stretch and the peak tension after the rapid stretch, at pCa 9 and at pCa 4, in the presence and absence of 30 mM BDM.
Statistics. Data are expressed as mean ± SE. Significant differences were assigned (P < 0.05) by using the paired or unpaired Student's t test as appropriate.
Results and Discussion
The human titin gene codes for conserved ≈28-residue PEVK repeats and E-rich motifs (12). We expressed fragment wt119-136, containing exons 119-136, with a central E-rich exon (no. 129) flanked by PEVK repeat exons (EMBL accession no. AJ277892; see Fig. 3A). Proteins were studied by atomic force microscope, and example force-extension curves for wt119-136 are shown in Fig. 1A. Force during stretch (blue) largely overlaps force during release (red) with little hysteresis. The initial part of the stretch curve was often irregular (Fig. 1 A Middle) but could also be smooth (Fig. 1 A Bottom). When present, these irregularities may result from interactions between the molecule and the substrate/cantilever tip (15). Accordingly, we focused our analysis on release curves, which were always smooth, fitted force-extension curves with the WLC equation (black lines in Fig. 1 A), and determined the PL, a measure of the molecule's bending rigidity (24).
Fig. 3.
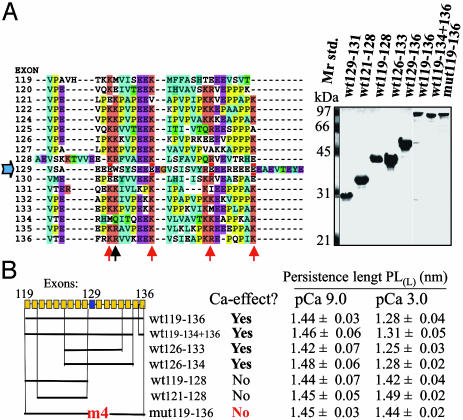
Calcium dependence of PL of deletion constructs and mutant naked PEVK fragments. (A) Alignment of amino acids (exon 123 omitted because it has not been found at the protein level). The central exon 129 is distinct from the flanking ≈28-residue PEVK repeats and is E-rich (18 E/38 residues). Exon 129 contains four glutamates at positions containing conserved lysines in neighboring exons (red arrows). These residues were mutated to lysines in mut119-136. Black arrow indicates tryptophane in exon 129 that was used for fluorescence spectroscopy (see Results and Discussion). (Right) SDS/PAGE of purified fragments. The numbers reflect represented exons. (B)PLL measured at low (pCa 9) and high (pCa 3) calcium. PL of fragments that contain both the E-rich domain and flanking PEVK repeats is significantly lower at high calcium. Calcium response is absent in mutant and fragments containing only PEVK repeats. Each value is the mean ± SEM of >100 force curves from six experiments.
Fig. 1.
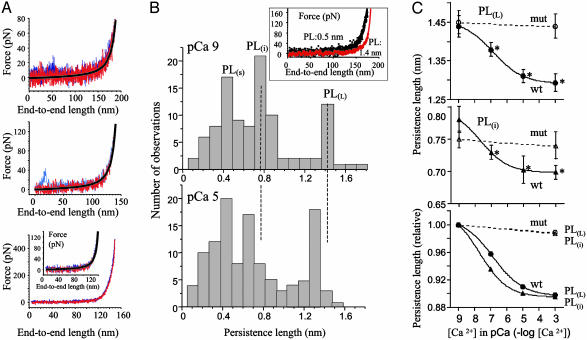
(A) Stretch (blue)-release (red) curves of wt119-136 (naked PEVK fragment) reveal that hysteresis is small both at low force (Top and Middle) and high force (Bottom; Inset shows expanded low force regime). Black lines indicate WLC curve fitted to release. (B) PL histograms are multimodal with three major peaks (S for short, i for intermediate, and L for long). Histograms show measurements in low calcium (pCa 9; Upper) and in presence of high free calcium (pCa 5; Lower). Note that calcium shifts peaks to shorter lengths. Cluster analysis revealed peaks at 0.44 nm, 0.78 nm, and 1.42 nm in low calcium and 0.43 nm, 0.70 nm, and 1.31 nm in high calcium. (Inset) Example of force-extension curve with long PL (red) and short PL (black). (C) PL varies with [Ca2+] in the pCa 9-5 regime (asterisks, significant difference vs. pCa 9). PLL (Top) and PLi (Middle) behave similarly. [PLS revealed a similar trend (not shown) but had only significantly reduced PL at the highest calcium level tested]. (Bottom) PL relative to values at pCa 9. Results with fragment mut119-136 (mut) are also displayed (see Results and Discussion). Each data point is the mean ± SEM of >100 force curves obtained from six experiments.
PL histograms were discrete, with peaks at ≈0.5, ≈0.7, and ≈1.4 nm (Fig. 1B). Although various explanations for the multimodal distributions exist, including the existence of multiple conformations (25), an explanation worth considering is titin's tendency to multimerize (26) and pulling either single molecules or multiple molecules tethered in parallel. The force required to produce a given fractional extension of n molecules tethered in parallel is n times that for a single molecule (24). Because, at a given fractional extension, force and PL are inversely proportional (24), the PL of n molecules will be that of the single molecule divided by n. We determined the PL histogram for eight different protein preparations (wt119-136). The PL of the peak at the longest (PLL), intermediate(PLi), and shortest lengths (PLS) were 1.44 ± 0.03 nm, 0.78 ± 0.02 nm, and 0.44 ± 0.02 nm, respectively. These values are within expected experimental error, assuming the peaks represent singlets (PL, 1.44 nm), doublets (PL, 1.44/2 nm), and triplets (PL, 1.44/3). These findings support the notion that the PL histogram allows the single molecule to be identified from the peak at the longest PL (PLL) and that peaks at shorter lengths reflect the PL of doublets (PLi) and triplets PLS).
Calcium left-shifted the histogram peaks (Fig. 1B). SAS cluster analysis confirmed the existence of three peaks in the histograms and established that, in the presence of calcium, the peaks were shifted to significantly shorter lengths. Experiments with different coating surfaces (mica, glass, gold-coated glass) gave identical results [except that with mica the histogram peak at the long length (PLL) was more prominent] and all revealed that calcium significantly reduced PL.
The half-maximal effect ([Ca]50) was observed at ≈0.15 μM (Fig. 1C). This concentration is within the physiological calcium range in muscle (≈0.05-10 μM). Magnesium had no effect (PLL, 1.45 ± 0.04 nm and 1.42 ± 0.03 nm at 0 and 4 mM Mg2+, respectively). Thus, the PEVK segment responds specifically to calcium, with a sensitivity that suggests a physiologically significant effect.
As an alternative to using naked PEVK fragments and identifying the single molecule PL from a histogram, we engineered fusion proteins comprising centrally located PEVK sequences with flanking Ig domains. During stretch, poly-Ig fragments reveal a “sawtooth-like” force curve, with each abrupt force drop reflecting unfolding of an individual domain; the regularity of the force peak separation and the magnitude of the unfolding forces can be used as a single molecule fingerprint (27, 28). We made two types of fusion proteins. One contains 235 PEVK residues (with the centrally located E-rich exon 129) flanked at its N terminus by the Ig domains I3-I5 and at its C terminus by I8-I10. The other contains a 485-residue PEVK insert (with the centrally located E-rich exon 129) flanked by I92-I94 and I95-97. We refer to these fragments as PEVKS and PEVKL (S for short and L for long), respectively (Fig. 2 A and B).
Fig. 2.
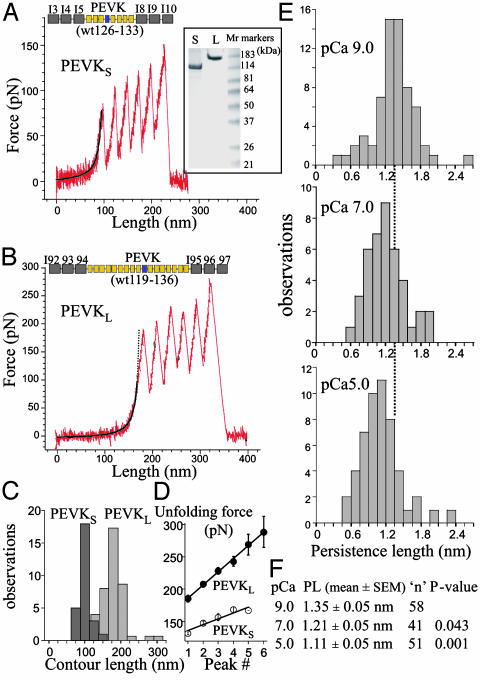
Atomic force microscope experiments with fragments PEVKS and PEVKL containing PEVK sequences flanked by Ig domains. Shown at the top of A and B are fragment composition: yellow, PEVK repeats; blue, E-rich exon; and gray, Ig domains. (Inset) SDS/PAGE of proteins used for atomic force microscopy (S, PEVKS; L, PEVKL). (A and B) Examples of force extension curves measured during stretch of PEVKS (A) and PEVKL (B). Initial part of force curve shows WLC fit (based on 0-50 pN force range): PL, 1.39 and 1.40 nm; CL, 102.8 and 182.9 nm for PEVKS and PEVKL, respectively. (C) Contour length histogram obtained by fitting the force extension curve before the first unfolding peak (using full force range) with WLC equation. (D) Unfolding force vs. peak number. (E) PL histogram of PEVKL curves with single molecule fingerprint (four or more force peaks during stretch, force peaks that are spaced by ≈28 nm, and unfolding forces that increase with peak number in the range of ≈200-300 pN). WLC fit was used on force-extension curve before first unfolding peak (0-50 pN force range). (F) Statistical analysis of data in E.
When these proteins are stretched, they initially develop low force. This result is followed by a steep force increase that leads to a sawtooth-like force extension curve (Fig. 2 A and B). The maximal number of force peaks before detachment of the molecule is six, and the peak separation (determined from WLC fits) is ≈28 nm, suggesting that each force drop reflects the unfolding of a single Ig domain (27, 28).
Observing that six unfolding peaks occurred infrequently (under our conditions, 1 in ≈20,000 “surface probings”), we therefore accepted curves with at least four regularly spaced force peaks for analysis, to ensure stretching the PEVK sequence. We determined the CL of the curve that leads up to the first force peak and found the CL of PEVKS to be significantly shorter than that of PEVKL (Fig. 2C). The obtained values are 98 ± 3 nm (n = 27) for PEVKS and 198 ± 4.0 nm (n = 61) for PEVKL. These values are similar to the CL of the PEVK regions contained in the fragments [PEVKS, 235 × 0.38 nm (= 89 nm); PEVKL, 485 × 0.38 nm (= 184 nm)], supporting that the force-extension curve before the first unfolding peak reflects extension of mainly the PEVK sequence.
The mean unfolding force (stretch velocity 0.5 μm/s) is higher for PEVKL (218 ± 3 pN; n = 156) than for PEVKS (145 ± 4 pN; n = 168). Furthermore, the slope of the unfolding force vs. peak number is significantly higher for PEVKL (Fig. 2D). These results are consistent with measurements on poly-Ig fragments with Ig domains from the distal and proximal tandem Ig segment (28). Thus, our results support that unfolding characteristics of Ig domains that are part of the PEVK-Ig fusion proteins may be used as a single molecule fingerprint whereas the force extension curve before the first Ig unfolding event reflects mainly PEVK segment extension.
We studied the effect of calcium on the PL of the PEVK segment, as reflected by the PL of the initial part of the stretch curve. Considering the infrequent occurrence of curves with the desired fingerprint, experiments were focused on only one of the fragments (PEVKL). We noted that performing a WLC fit on the full force-extension curve before the first unfolding peak resulted in a consistent error term at intermediate and high force levels (>≈100 pN). Whether this results from unfolding intermediates that have been reported for Ig domains at force levels of ≈100 pN (29), or from another source, remains to be established. Fitting with a modified WLC equation that contains an enthalpic term or by using the freely jointed chain model (17) did not significantly improve the fit. We therefore used the basic WLC chain model and restricted the force range that was used to 0-50 pN (similar to that used for the naked PEVK fragment). The WLC fitted these data well, with error terms that were scattered around zero. Results indicate that increasing [Ca2+] results in a small but statistically significant PL reduction (see Fig. 2 E and F). A reduction was seen when free [Ca2+] was increased from pCa 9 to pCa 7, and the reduction was enhanced when calcium was further increased to pCa 5.0. In conclusion, experiments with naked PEVK fragments and PEVK-Ig fusion proteins both support that calcium reduces the PEVK PL.
To identify the region(s) responsible for the calcium effect, we studied several deletion constructs (Fig. 3). We focused on naked PEVK fragments because these can be studied by fluorescence (see below) without confounding effects due to Ig domain sequences. An advantage is also that characterizing naked PEVK fragments can be achieved with relatively high throughput (unlike work with PEVK-Ig fusion proteins). The minimal fragment that responds to calcium contains a central E-rich domain flanked by three PEVK repeats (Fig. 3B). Fragments comprising solely PEVK repeats do not respond to calcium: inclusion of the E-rich domain is required. Because the E-rich domain is too small (38 residues) to measure only its force-extension curve, we studied whether the calcium effect is derived from this domain as follows. Sequence alignment revealed four highly conserved lysine residues in the ≈28-residue PEVK repeat family (Fig. 3A, red arrows) that in exon 129 are replaced by glutamates. To test whether these glutamates are involved in the calcium response we converted them to lysine residues by using site-directed mutagenesis. The PL of the mutant protein (mut119-136) was calcium insensitive (Figs. 1C and 3B). Thus, the calcium effect requires the presence of glutamates in the E-rich motif.
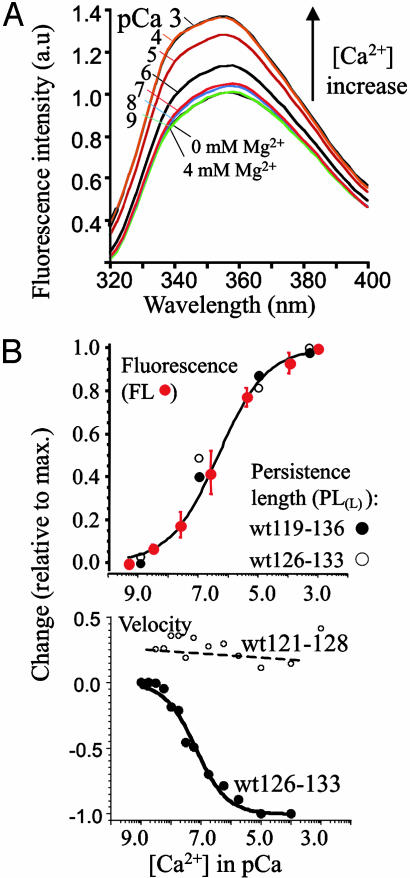
The effect of calcium on the conformation of the PEVK segment was examined by using fluorescence spectroscopy, focusing on the smallest fragment that responded to calcium (wt126-133) because it contains a single tryptophane (Fig. 3A, black arrow) near the four critical glutamates (above) and because high molar concentrations of soluble protein can be obtained. The emission spectrum was insensitive to magnesium but responded to calcium with increased intensity at λmax (Fig. 4A) with a [Ca]50 of ≈0.4 μM (Fig. 4B). Dialyzing the proteins against calcium-free solutions and repeating the fluorescence experiments allowed us to reproduce the calcium effect, indicating that the underlying changes in the PEVK segment are reversible. However, the calcium effect was absent when protein degradation had occurred, and mass spectrometry revealed that the full-length fragment (wt126-133) was required for the emission spectrum to respond to calcium. Fig. 4B Upper shows that the PL of fragments wt126-133 and wt119-136 vary with calcium in a manner similar to that observed for fluorescence intensity.
Fig. 4.
Calcium effect on PEVK structure. (A) Fluorescence emission spectrum of wt126-133. Numbers indicate [Ca2+] in pCa. Intensity increases with [Ca2+] between pCa 9 and 4 and then saturates (curves at pCa 4 and 3 exactly overlap). Two bottom curves (green and black) also overlap: they were in 0 mM (green) and 4 mM (black) magnesium. (B Upper) Calcium-dependent fluorescence (FL) intensity change at λmax. (Mean values ± SEM of three experiments.) Also shown are PL values (PLL) of wt119-136 (filled symbols) and wt126-133 (open circles). (Each data point is the mean of >100 force curves obtained from six experiments.) (Lower) Calcium dependence of ultrasonic velocity of solutions containing wt126-133 (PEVK repeats and E-rich exon) and wt121-128 (PEVK repeats only). Change in velocity is shown relative to the maximal change observed for wt126-133 between pCa 8.5 and 5.0 (velocity change of -16.7 cm/s). Note a steep and significant decrease in velocity only for wt126-133.
Because PEVK repeats are devoid of tryptophanes and tyrosines (Fig. 3A), they could not be studied with fluorescence, and we therefore used ultrasonic velocimetry (see Methods)asan independent assay for calcium-induced changes in fragment wt121-128 (PEVK repeats only) and wt126-133 (containing PEVK repeats and the E-rich exon). We found that wt126-133 responded to calcium by decreasing the ultrasonic velocity (Fig. 4B Lower) with a half maximal effect at ≈0.07 μM [Ca]50. In contrast, calcium did not significantly affect the ultrasonic velocity of fragment wt121-128. Overall, these results support the idea that calcium affects the conformation of the PEVK segment, that this requires the presence of the E-rich exon 129, and that calcium sensitivity is high with a half-maximal effect in the submicromolar range. This view is consistent with the 0.1 μM Kd reported by Tatsumi et al. (14) for binding of calcium to a PEVK-containing degradation product of native titin. Thus, E-rich motifs are likely to bind calcium with high affinity and cause conformational (increase in fluorescence, decrease in ultrasonic velocity) and mechanical (decrease in PL) changes in titin's PEVK segment.
We can only speculate about the mechanism(s) involved. Perhaps the E-rich exon and flanking sequences form in absence of calcium an ordered structure that on calcium binding becomes disordered, giving rise to a more flexible chain with a lower PL. Alternatively, the E-rich exon may be disordered in the absence of calcium and negatively charged glutamates give rise to a relatively stiff chain, due to electrostatic repulsion between these charges. Calcium binding to glutamates may then reduce the “electrostatic stiffening effect” and increase the flexibility of the chain (i.e., reduce the PL). Future high-resolution structural studies (NMR, crystallography) in the presence and absence of calcium may establish the mechanism(s) underlying our findings.
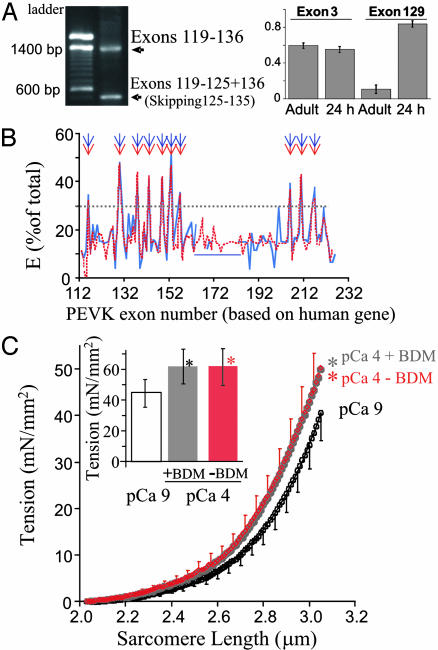
In human titin, differential splicing events control the PEVK segment composition, with N2B cardiac titin containing solely PEVK repeats and N2BA cardiac and skeletal muscle titin isoforms containing a variable number of PEVK repeats and E-rich motifs (12). During amplification of exons 119-136 from human skeletal muscle by RT-PCR, we noted consistently the amplification of a smaller product(see Fig. 5A Left). Sequencing showed that this is a splice variant that excludes the E-rich exon 129. Similarly, the E-rich exon 129 is present in rabbit psoas muscle, but at much higher levels in neonates than in adults (Fig. 5A Right), indicating that the splicing of exon 129 is developmentally regulated. It seems that E-rich motifs are under differential splicing control, suggesting a role that may be required in some cells but that must be avoided in others. The human titin gene contains 10 E-rich motifs, all of which are conserved in the mouse gene (Fig. 5B). Comparison of the human and mouse amino acid sequence of E-rich exons reveals high homology (93% for exon 129), again suggesting that these sequences have important biological functions, and it will be important to test whether all E-rich exons are calcium sensitive. The idea that E-rich motifs perform important functions is further supported by the recent finding of a Caenorhabditis elegans titin with a PEVK region that also contains E-rich motifs that interrupt tandemly arranged repeats (11).
Fig. 5.
(A Left) PCR with exon119-sense plus exon136-reverse primers and human skeletal muscle cDNA results in not only a full-length product but also a 500-bp product in which exons 126-135 are spliced out (right lane, bottom). (Right) Hybridization of rabbit psoas muscle cDNAs to probes specific to titin exons 129 and 3 reveals developmental regulation of exon 129 expression, with low levels in adult animals and ≈8-fold higher levels in 24-h-old neonates (for technical details, see Supporting Text). (B) Glutamate content in PEVK exons of human (red) and mouse (blue) titin gene (curves overlap). Human has 114 PEVK exons, 10 of which have >30% glutamate (E) content. All E-rich motifs are conserved in human (red arrows) and mouse (blue arrows) genes. (C) Calcium effect on tension developed by muscle fibers from which actomyosin interaction had been abolished (see Results and Discussion). Relation between tension and SL measured during slow stretch (0.1 μm per half sarcomere sec) at pCa 9 (open circles) and pCa 4 with (gray symbols) and without (red symbols) 30 mM BDM (pCa 4 data sets overlap). (Inset) Tension at end of rapid stretch (10 μm per half sarcomere sec) from slack length to SL 2.7 μm. Calcium addition resulted in a BDM-independent tension increase. Shown are mean values ± SEM of six fibers. *, Significant difference between pCa 4 and pCa 9 results.
Of the 10 E-rich motifs, 9 (including exon 129) can be identified in soleus cDNA (1), and we tested whether titin-based tension of soleus fibers is calcium dependent. Mouse skinned fibers were used in which actomyosin interaction was completely abolished by extracting thin filaments with gelsolin (see Methods). Tension was measured during a slow stretch to a SL of ≈3.1 μm, as well as after a rapid stretch to a SL of 2.7 μm. It is well established that forces of mechanically skinned fibers in the absence of calcium (pCa 9) are largely titin-based. Adding to these thin-filament extracted fibers 100 μM free Ca2+ (pCa 4) significantly increased tension during a slow stretch (Fig. 5C, red symbols), as well as at the peak after a rapid stretch (Fig. 5C Inset, red bar). Because actomyosin-based tension at short SL was undetectable in these fibers, and the increase in calcium-induced tension on stretch was insensitive to the actomyosin inhibitor BDM (Fig. 5C), it is unlikely that residual actomyosin interaction explains these findings. Rather, the tension increase in calcium is likely due to titin. This conclusion is consistent with the calcium-induced PL decrease of PEVK molecules [force at a given fractional extension is inversely proportional to PL (24)]. Thus, the finding that calcium increases titin-based tension of muscle fibers is supported by studies at the level of the single molecule.
Titin's calcium-sensitive tension may explain the calcium-induced tension of sarcomeres stretched beyond thin and thick filament overlap (30), a phenomenon that cannot be explained by the sliding filament/crossbridge model of muscle contraction (31). Furthermore, many investigators have shown that, when a stretch is imposed on a contracting muscle (as occurs during normal activity), the ensuing force response cannot be explained solely by actomyosin interaction, and the existence of a calcium-sensitive parallel elastic element has been postulated (23, 32, 33). Pioneering work by Bagni et al. (23, 34) revealed a “static tension” in intact fibers with a time course similar to that of intracellular free calcium and a magnitude that is unaffected by BDM. The static tension increases with stretch amplitude and starting SL, reaching ≈6% of maximal actomyosin tension at an SL of 2.1 μm and is several fold higher at 2.8 μm SL. Bagni et al. (23) suggested a mechanism based on titin. Campbell and Moss (32) studied rat soleus fibers and concluded that the initial tension and stiffness of a non-crossbridge structure are severalfold greater in pCa 4.5 solution than pCa 9.0 solution. These authors postulate the existence of a calcium-sensitive parallel elastic element. Our work indicates that such an element indeed exists and that calcium sensitivity is derived from E-rich PEVK motifs in titin. Thus, when a muscle is activated, the rise in intracellular calcium not only triggers actomyosin-based tension but also additional titin-based tension. This calcium-sensitive tension of titin increases with SL, in contrast to actomyosin-based tension that decreases with SL, and may play important roles in stabilizing muscle during contraction.
Supplementary Material
Acknowledgments
We thank Dr. L. Gloss for help with fluorescence, Dr. M. Gotthardt for sequence analysis, Drs. K. Campbell and B. Slinker for reading of the manuscript, and Mrs. H. Zhou for assistance. We thank Asylum Research (Santa Barbara, CA) for custom-written software. This work was supported by Grants HL61497 and HL62881 from the Heart Lung and Blood Institute of the National Institutes of Health and La668/6-2/7-1 from the Deutsche Forschungsgemeinschaft.
Abbreviations: PEVK, proline-glutamate-valine-lysine; CL, contour length; PL, persistence length; SL, sarcomere length; WLC, worm-like chain; BDM, 2,3-butanedione monoxime.
References
- 1.Labeit, S. & Kolmerer, B. (1995) Science 270, 293-296. [DOI] [PubMed] [Google Scholar]
- 2.Tskhovrebova, L. & Trinick, J. (2002) Philos. Trans. R. Soc. London B Biol. Sci. 357, 199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granzier, H. & Labeit, S. (2002) J. Physiol. London 541, 335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maruyama, K. (1997) FASEB J. 11, 341-345. [DOI] [PubMed] [Google Scholar]
- 5.Wang, K. (1996) Adv. Biophys. 33, 123-134. [PubMed] [Google Scholar]
- 6.Trombitas, K., Greaser, M., Labeit, S., Jin, J. P., Kellermayer, M., Helmes, M. & Granzier, H. (1998) J. Cell Biol. 140, 853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linke, W. A., Ivemeyer, M., Mundel, P., Stockmeier, M. R. & Kolmerer, B. (1998) Proc. Natl. Acad. Sci. USA 95, 8052-8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuzawa, A., Shimamura, J., Takemori, S., Kanzawa, N., Yamaguchi, M., Sun, P., Maruyama, K. & Kimura, S. (2001) EMBO J. 20, 4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Champagne, M. B., Edwards, K. A., Erickson, H. P. & Kiehart, D. P. (2000) J. Mol. Biol. 300, 759-777. [DOI] [PubMed] [Google Scholar]
- 10.Southgate, R. & Ayme-Southgate, A. (2001) J. Mol. Biol. 313, 1035-1043. [DOI] [PubMed] [Google Scholar]
- 11.Flaherty, D. B., Gernert, K. M., Shmeleva, N., Tang, X., Mercer, K. B., Borodovsky, M. & Benian, G. M. (2002) J. Mol. Biol. 323, 533-549. [DOI] [PubMed] [Google Scholar]
- 12.Bang, M. L., Centner, T., Fornoff, F., Geach, A. J., Gotthardt, M., McNabb, M., Witt, C. C., Labeit, D., Gregorio, C. C., Granzier, H. & Labeit, S. (2001) Circ. Res. 89, 1065-1072. [DOI] [PubMed] [Google Scholar]
- 13.Greaser, M. (2001) Proteins 43, 145-149. [DOI] [PubMed] [Google Scholar]
- 14.Tatsumi, R., Maeda, K., Hattori, A. & Takahashi, K. (2001) J Muscle Res. Cell Motil. 22, 149-162. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe, K., Nair, P., Labeit, D., Kellermayer, M. S., Greaser, M., Labeit, S. & Granzier, H. (2002) J. Biol. Chem. 277, 11549-11558. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe, K., Muhle-Goll, C., Kellermayer, M. S., Labeit, S. & Granzier, H. (2002) J. Struct. Biol. 137, 248-258. [DOI] [PubMed] [Google Scholar]
- 17.Bustamante, C., Marko, J. F., Siggia, E. D. & Smith, S. (1994) Science 265, 1599-1600. [DOI] [PubMed] [Google Scholar]
- 18.Granzier, H. L. & Wang, K. (1993) Biophys. J. 65, 2141-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taulier, N. & Chalikian, T. V. (2001) J. Mol. Biol. 314, 873-889. [DOI] [PubMed] [Google Scholar]
- 20.Granzier, H. L. & Irving, T. C. (1995) Biophys. J. 68, 1027-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granzier, H., Helmes, M. & Trombitas, K. (1996) Biophys. J. 70, 430-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granzier, H., Kellermayer, M., Helmes, M. & Trombitas, K. (1997) Biophys. J. 73, 2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagni, M. A., Cecchi, G., Colombini, B. & Colomo, F. (2002) Biophys. J. 82, 3118-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellermayer, M. S., Smith, S. B., Granzier, H. L. & Bustamante, C. (1997) Science 276, 1112-1116. [DOI] [PubMed] [Google Scholar]
- 25.Li, H., Oberhauser, A. F., Redick, S. D., Carrion-Vazquez, M., Erickson, H. P. & Fernandez, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 10682-10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tskhovrebova, L. & Trinick, J. (1997) J. Mol. Biol. 265, 100-106. [DOI] [PubMed] [Google Scholar]
- 27.Carrion-Vazquez, M., Oberhauser, A. F., Fisher, T. E., Marszalek, P. E., Li, H. & Fernandez, J. M. (2000) Prog. Biophys. Mol. Biol. 74, 63-91. [DOI] [PubMed] [Google Scholar]
- 28.Li, H., Linke, W. A., Oberhauser, A. F., Carrion-Vazquez, M., Kerkvliet, J. G., Lu, H., Marszalek, P. E. & Fernandez, J. M. (2002) Nature 418, 998-1002. [DOI] [PubMed] [Google Scholar]
- 29.Marszalek, P. E., Lu, H., Li, H., Carrion-Vazquez, M., Oberhauser, A. F., Schulten, K. & Fernandez, J. M. (1999) Nature 402, 100-103. [DOI] [PubMed] [Google Scholar]
- 30.Pollack, G. H. (1988) J. Mol. Cell. Cardiol. 20, 563-570. [DOI] [PubMed] [Google Scholar]
- 31.Huxley, H. E. (1969) Science 164, 1356-1365.4181952 [Google Scholar]
- 32.Campbell, K. S. & Moss, R. L. (2002) Biophys. J. 82, 929-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herzog, W. & Leonard, T. R. (2002) J. Exp. Biol. 205, 1275-1283. [DOI] [PubMed] [Google Scholar]
- 34.Bagni, M. A., Cecchi, G., Colomo, F. & Garzella, P. (1994) J. Physiol. (London) 481, 273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.