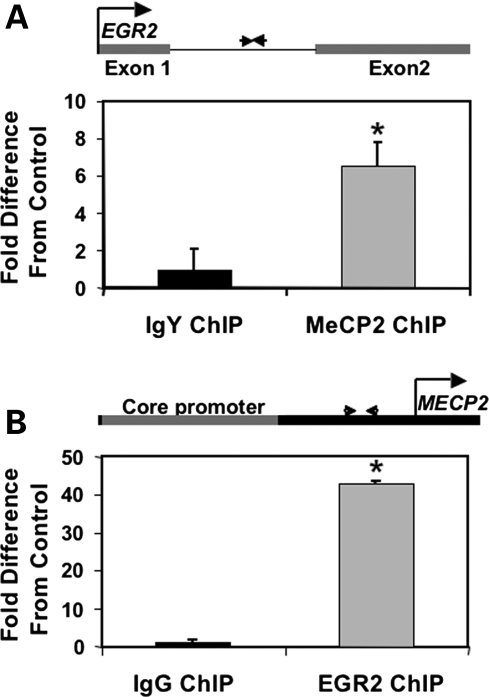
Figure 1.
(A) ChIP using anti-MeCP2 or non-specific IgY was performed on chromatin from PMA-stimulated human neuroblastoma cells in two separate experiments. qPCR was performed using primers designed to a DNA sequence in the EGR2 intron which contains a putative MAR and CpGs in the vicinity of A/T runs (diagrammed above). MeCP2 ChIP was significantly enriched compared with the IgY Control ChIP normalized to one (*P < 0.03 by Wilcoxon). Results are the mean ± SEM of six replicates. (B) ChIP was performed using anti-EGR2 or non-specific IgG and primers were designed to a region between the MECP2 core promoter and transcriptional start site which contained a predicted EGR2-binding site (diagrammed above). EGR2 ChIP was significantly enriched compared with the RIgG Control ChIP normalized to 1 (*P < 0.02 by Wilcoxon). Results are the mean ± SEM of seven replicates. Both the experimental and the Control ChIP experiments were normalized to input.