Figure 2.
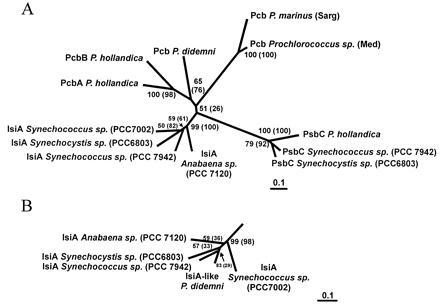
Phylogenetic distance tree (neighbor-joining method; ref. 23) of Pcb, IsiA, and PsbC proteins from prochlorophytes and cyanobacteria. Numbers at the branch points represent the bootstrap values for 100 replicate trees; numbers in parentheses are the bootstrap values for the parsimony analysis that generated a similar tree (not shown). The protein sequences were aligned using clustal v (19); trees were generated using protdist and neighbor (pam matrix) or protpars programs of the phylip package (20). (A) Tree using 308 common residues. All regions with gaps were excluded from the analysis, but the same configuration of branches resulted when gaps were included. (B) IsiA branch from trees that included the IsiA-like sequence from P. didemni, using the 206 common residues.
