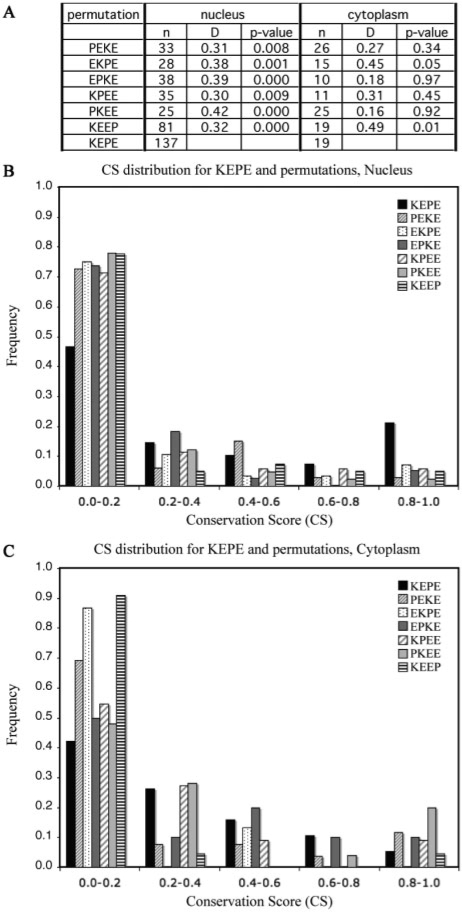
Fig. 2.
Conservation score distributions for the KEPE motif and the six permutations comparing the nuclear and cytoplasmic compartments. KEPE bearing proteins were retrieved from UniProt/Swiss-Prot with the compartment keyword expressions in Table 1, processed for IUPred disorder prediction and evaluated for the ELM CS score. To enable comparison, the sets have been normalized into percentages and sorted into five CS score bins. The table in (A) shows that the KEPE matches have a significantly different conservation distribution in the nucleus compared with the controls. n=number of instances, D=maximum difference between the cumulative distributions, P-value=significance of the difference D, according to the KS test. KEPE matches also show a peak of strong conservation (unmatched by the controls) in the nucleus (B) but not in the cytoplasm (C). These results are consistent with a lack of functionality of the KEPE motif in the cytosol (since this is the implied meaning of ‘cytoplasm’ in Swiss-Prot).