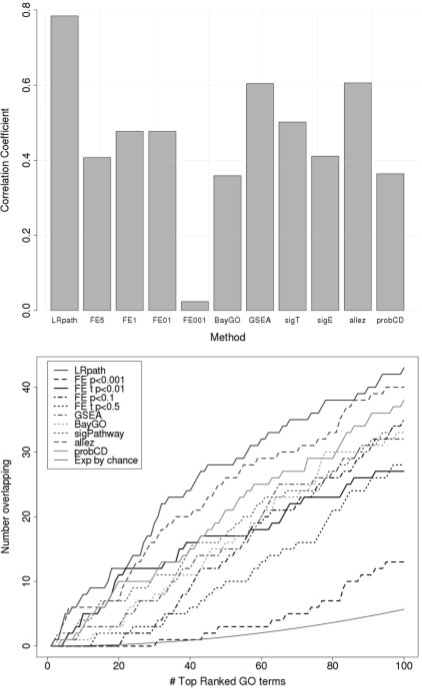
Fig. 3.
Concordance of methods between two independent Breast Cancer datasets. Reproducibility of the methods (LRpath, FE with cutoffs of 0.50, 0.10, 0.01 and 0.001 for DEGs, BayGO, GSEA, sigPathway, allez and ProbCD, respectively) was tested by measuring the consistency of results across two datasets, both comparing grade 3 to grade 1 tumors. (A)Correlation between datasets for each method. As a measure of significance, the –log(P-values) of GO term enrichment were calculated for each method and dataset separately, and the Pearson correlation coefficients between datasets were calculated. (B) Overlapping enriched GO terms by rank. Ranked lists of GO terms were generated for each method and each dataset separately. The number of overlapping GO terms was calculated between datasets for each method for increasing length of ranked lists.