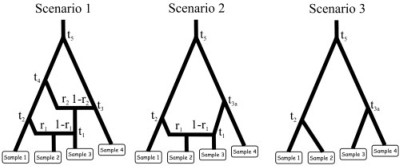
Fig. 1.
First example: the three evolutionary scenarios. The dataset used as an example has been simulated according to scenario 1 (left). The parameter values were the following: all populations had an effective (diploid) size of 1000, the times of successive events (backward in time) were t1 = 10, t2 = 500, t3 = 10 000, t4 = 20 000 and t5 = 200 000, the two admixture rates were r1 = 0.6 and r2 = 0.4. Scenario 1 includes six populations, the four that have been sampled and two left parental populations in the admixture events. Scenario 2 and 3 include 5 and 4 populations, respectively. Samples 3 and 4 have been collected 2 and 4 generations earlier than the first two samples, hence their slightly upward locations on the graphs. Time is not at scale.