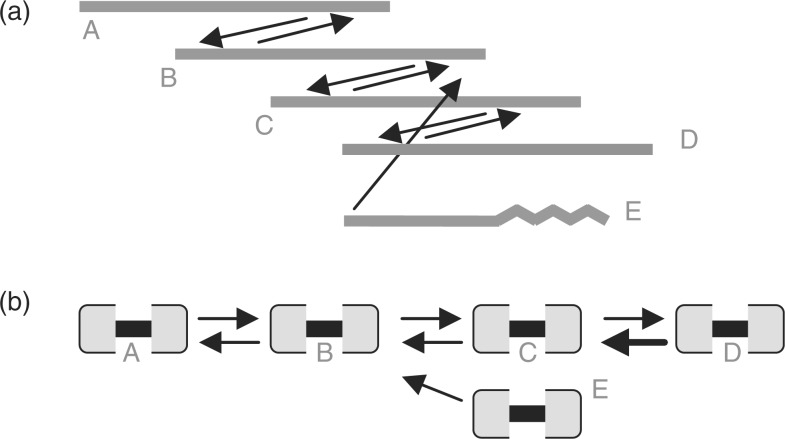
Fig. 1.
Two representations of a best overlap graph. In (a), the layout resembles a multiple sequence alignment. In (b) each read is represented by two nodes joined by an undirected edge. Arrows represent best overlaps, where best means covering the most sequence. There are mutual best overlaps between successive pairs of reads A through D. Due to erroneous bases at one end (wavy line), read E has a non-mutual best overlap to B. Paths span undirected and directed edges alternately. Path EBA converges on path ABCD. CABOG scores read E lower than the others since only three reads are on paths from it. Starting with any one of the high-scoring reads, CABOG would build initial unitig ABCD, then E. Using saved information about each path intersection, CABOG would discount the intersection at B because the path from E spanned only one read before B. It would break ABCD only if there were also a change in read arrival rate at B, which is not the case here. Although linear-time directed-path following finds the longest possible unitig in this constructed case, it is not guaranteed to do so when paths span multiple intersections.