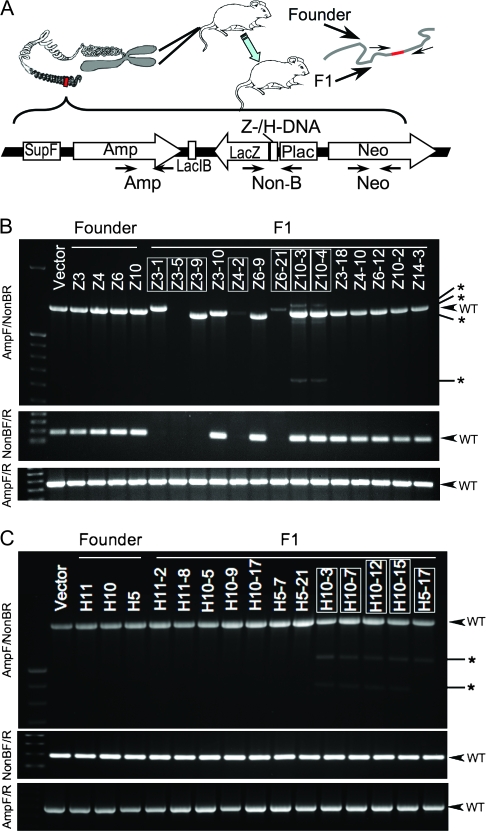
Figure 1.
DNA secondary structure-induced genetic instability in mouse chromosomes. A) Schematic structure of the linearized mutation reporter shuttle plasmid p2RT (constructed based on plasmid pUC19, Invitrogen, Carlsbad, CA), which was integrated in the FVB/N mouse genome by microinjection into fertilized oocytes. Primers used for genotyping are also shown in the figure. Amp = ampicillin resistance; Neo = neomycin resistance; Non-B = inserted H-DNA, Z-DNA, or control sequence. For sequences of primers, H-DNA, Z-DNA, or control sequence and the context sequence in the vector, see Supplementary Figure and Table (available online). B) Polymerase chain reaction (PCR) analysis of the p2RT-CG14 (Z-DNA) mouse strain. C) PCR analysis of the p2RT-MYC (H-DNA) mouse strain. Three primer pairs shown in panel (A) were used to amplify the genomic DNA from tail samples of each F1 mouse. Top: AmpF/Non-BR primers; middle: NonBF/R primers; bottom: AmpF/R primers. Samples from mice demonstrating DNA secondary structure-induced genetic instability are boxed. The expected PCR products from wild-type (WT) transgenes are indicated by arrows; abnormal products are indicated by asterisks.