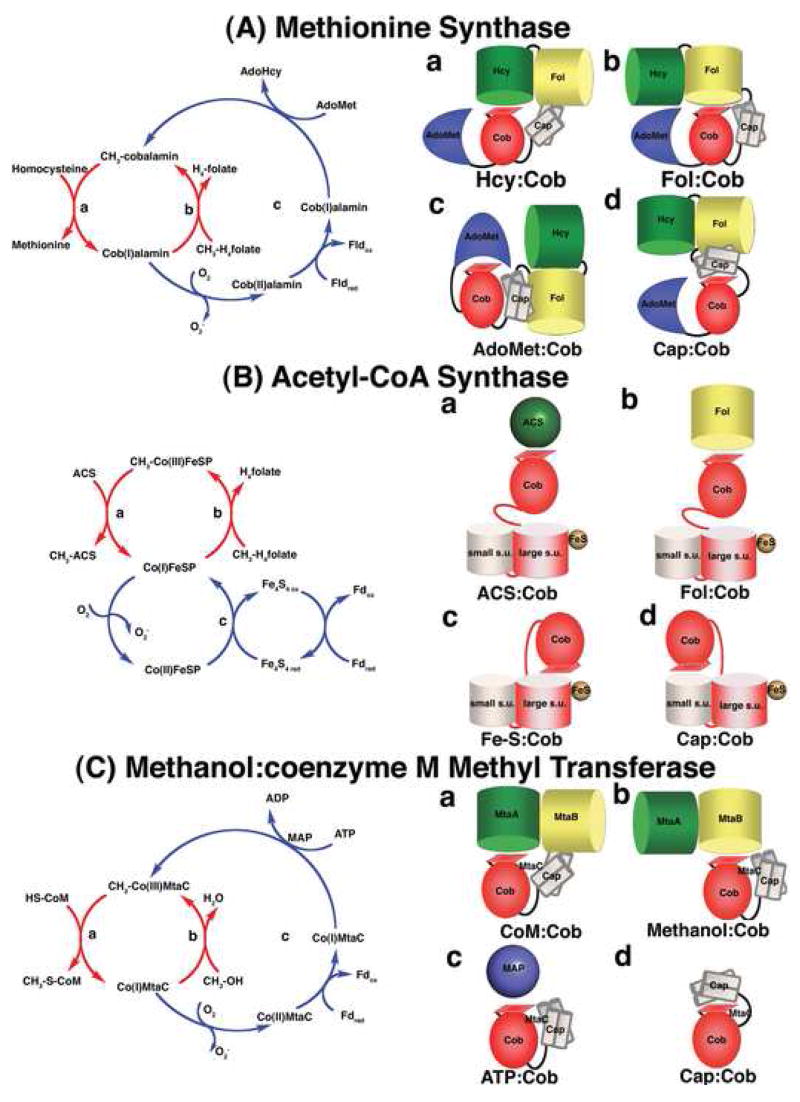
Figure 2. The catalytic cycles and proposed conformations of three well-studied methyltransferases.
(A) Methionine synthase The cobalamin cofactor is alternately methylated by CH3-H4folate bound to the Fol domain (b, requiring the Fol:Cob conformation cartooned on the right) and demethylated by homocysteine bound to the Hcy domain (a, Hcy:Cob). Occasionally, the cob(I)alamin cofactor undergoes oxidation. Return of the inactive cob(II)alamin species to the catalytic cycle require a reductive reactivation, in which adenosylmethionine bound to the C-terminal domain (c, AdoMet:Cob) serves as the methyl donor [7]. In E. coli, flavodoxin serves as the source of electrons for reductive activation [35], while in humans the source of electrons is methionine synthase reductase, a protein with homology to both flavodoxin and NADPH-ferredoxin (flavodoxin) oxidoreductase [38,39]. The remaining conformation (d, Cap:Cob) has only been seen in crystals of the isolated cobalamin-binding module. While the cartoon shows all four domains, x-ray structures of the full length protein have never been obtained, and the cartoons are based on docking of the N-terminal and C-terminal halves of the protein in a plausible manner. (B) Acetyl-CoA synthase. The 5-hydroxymethylbenzimidazolyl cobamide cofactor of the corrinoid iron/sulfur protein (AcsCD) is alternately methylated by CH3-H4folate bound to the AcsE methyltransferase (b, Fol:Cob) and demethylated by transfer of the methyl group to Ni+1 in acetyl CoA synthase (a, ACS:Cob). When the cob(I)amide undergoes occasional oxidation to cob(II)amide, it is returned to the catalytic cycle by reduction by the Fe4-S4 cluster on the large subunit (AcsC) of the corrinoid iron/sulfur protein (c, Fe-S:Cob). This cluster is in turn re-reduced with electrons derived from ferredoxin or from enzymes coupled to ferredoxin. The remaining conformation (d, Cap:Cob) is the one seen in the isolated corrinoid iron-sulfur protein [18••]). (C) Methanol:coenzyme M methyltransferase. The corrinoid-binding protein MtaC forms a complex with MtaA (the coenzyme M-binding methyltransferase) and MtaB (the methanol-binding methyltransferase). During catalysis the complex cycles between CoM:Cob (a) and Methanol:Cob (b) conformations. The latter complex has been characterized crystallo-graphically in the absence of MtaA. Activation of the cob(II)amide cofactor requires reduction by ferredoxin and is coupled to ATP hydrolysis by the action of the MAP protein (c, ATP:Cob). The Cap:Cob conformation (d) is hypothetical.