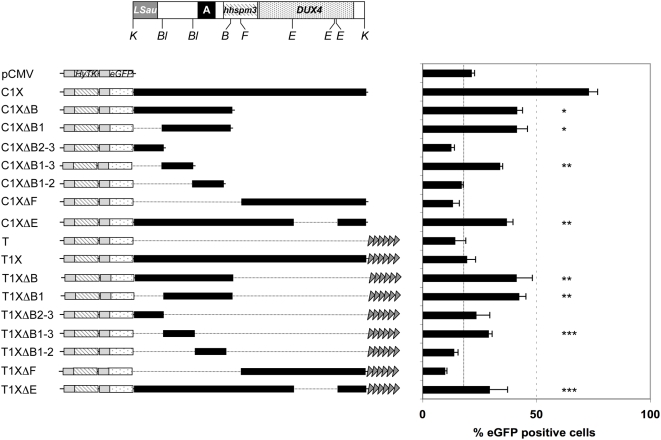
Figure 2. Mapping of the regulatory fragments within D4Z4.
A. Schematic representation of the D4Z4 element from position 1 to 3303 given relative to the two flanking KpnI sites (K) (to scale). The different regions within D4Z4 are indicated: LSau repeat (position 1–340), Region A (position 869–1071), hhspm3 (position 1313–1780), DUX4 ORF (position 1792–3063). The different restriction sites used for the cloning of D4Z4 subfragments are indicated (B: BamHI; Bl: BlpI; F: FseI; E: EheI). B. Different fragments obtained after digestion of D4Z4 were cloned downstream of the eGFP reporter (“C” constructs) or between the reporter gene and the telomeric seed (“T” constructs). Linearized plasmids were transfected into C33A cells and the percentage of eGFP positive cells was monitored by flow cytometry for an extended period of time. The histogram represents the mean value of the percentage of eGFP positive cells from day 18 to day 29 when eGFP expression reaches a plateau±S.D shown by error bars. Fragments DB2-3 (position 1 to 382), DB1-2 (position 814 to 1381) and DF (position 1549 to 3303) do not abrogate TPE or CPE while fragment DB1 (position 1 to 1381), DB1-3 (position 382 to 814) and DE (deleted of a distal 623 bp fragment from position 2269 to 2892) protect from CPE and TPE. Asterisks denote statistically significant values relative to control vectors (pCMV or T) (Student's t test). * p<0.001; **p<0.005; *** p<0.05.