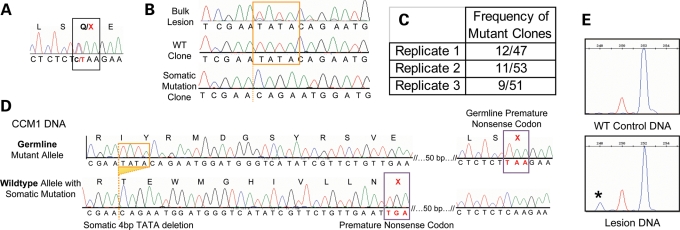
Figure 1.
A biallelic somatic mutation identified in CCM1 sample 2049. (A) Direct sequence analysis of bulk lesion tissue identified the germline mutation as the common Hispanic mutation, c.1363C>T, Q455X in exon 10 of CCM1. (B) Clonal analysis of CCM1 exon 10 reveals wild-type clones, germline mutant clones and clones with the 4 bp deletion somatic mutation, c.1270_1273DelTATA. The deleted bases are boxed in orange in the wild-type sequences, and the deletion site is marked with an orange dotted line in the somatic mutant sequence (bottom tracing). The somatic mutation is barely visible from direct sequence of bulk lesion. (C) The somatic mutation was identified in the lesion tissue multiple times in each of three rounds of PCR, cloning and sequencing. Replicates 1 and 2 used HiFi Platinum Taq Polymerase (Invitrogen) and replicate 3 used HotStar Polymerase (Qiagen). (D) The somatic and germline mutations are biallelic. Sequencing analysis of individual colonies containing both mutation loci shows three classes of clones, germline mutant alleles, wild-type alleles, and alleles carrying the somatic mutation that are wild-type for the germline mutation. (E) Fragment analysis validates the somatic mutation in the lesion. Normal control DNA shows all amplicons of predicted 252 bp length (blue peak). The red size marker is at 250 bp, and the lesion-derived DNA shows products of WT length and those from the somatic mutant allele at 248 bp. The mutant allele represents 11% of the total amplicons.