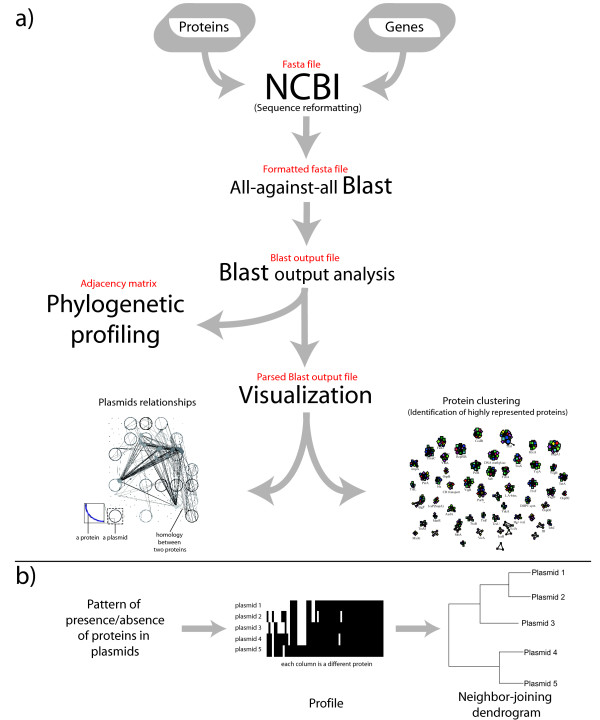
Figure 1.
B2N workflow and analysis. a) Scheme of the data workflow in B2N. The visualization is realized using Visone software. The input of each module (i.e. the output of the previous one) is shown in red fonts. b) Phylogenetic profiling of molecules in the dataset. Using the matrix of occurrence patterns, groups of proteins are identified at different threshold values. A new matrix is obtained composed of a row for each plasmid in the dataset and a number of columns corresponding to the number of groups in the network. Each entry i, j of the matrix contains 1 if at least one protein from plasmid i is present in cluster j, 0 if no protein from plasmid i is present cluster j. This matrix is used for calculating distances using the Jaccard metric and dendrogram construction. This analysis identifies those plasmids that contain similar proteins. By applying the same workflow in the second dimension of the phylogenetic profiles matrix, it is possible to find those protein clusters having similar occurrence patterns.