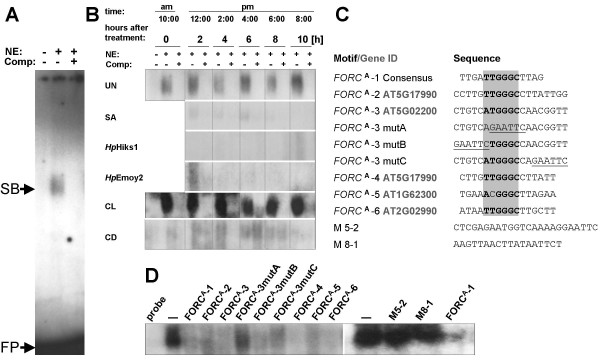
Figure 1.
In vitro-interactions of FORCA with nuclear Arabidopsis proteins. A, B: Interactions of FORCA with nuclear protein extracts from Arabidopsis Col-0 seedlings grown under normal light conditions and left untreated (UN) or treated with salicylic acid (SA), infected with the avirulent Hp isolates Hiks1 and Emoy2 or exposed for three days to continuous light (CL) or continuous darkness (CD). Panel (A) shows the whole gel for the 0 h untreated samples; panel (B) shows gel sections with the FORCA-specific band shift for all tested conditions. NE: nuclear extract, Comp: unlabeled competitor probe, SB: FORCA-specific band shift, FP: free probe.C: FORCA derived oligonucleotides used in EMSAs. The gene IDs specify the promoters from which each motif is derived. Positions representing the defining T/ATGGGC core sequences of the shown wild type FORCA permutations are highlighted by a grey box. Sequences mutated in the tested FORCA-3 permutations are underlined. D: EMSA competition analysis of FORCA-1 interactions using Arabidopsis nuclear protein extracts from Arabidopsis Col-0 seedlings exposed to CL.