Abstract
In birds and mammals T cells develop along two discrete pathways characterized by expression of either the αβ or the γδ T-cell antigen receptors (TCRs). To gain further insight into the evolutionary significance of the γδ T-cell lineage, the present studies sought to define the chicken TCRγ locus. A splenic cDNA library was screened with two polymerase chain reaction products obtained from genomic DNA using primers for highly conserved regions of TCR and immunoglobulin genes. This strategy yielded cDNA clones with characteristics of mammalian TCR γ chains, including canonical residues considered important for proper folding and stability. Northern blot analysis with the TCRγ cDNA probe revealed 1.9-kb transcripts in the thymus, spleen, and a γδ T-cell line, but not in B or αβ T-cell lines. Three multimember Vγ subfamilies, three Jγ gene segments, and a single constant region Cγ gene were identified in the avian TCRγ locus. Members of each of the three Vγ subfamilies were found to undergo rearrangement in parallel during the first wave of thymocyte development. TCRγ repertoire diversification was initiated on embryonic day 10 by an apparently random pattern of V-Jγ recombination, nuclease activity, and P- and N-nucleotide additions to generate a diverse repertoire of avian TCRγ genes early in ontogeny.
Keywords: chicken, evolution, ontogeny
Studies in the chicken, Gallus gallus domesticus, suggest that avian T-cell differentiation and function are similar to those described in mammals (1, 2). Divergent pathways of T-cell development are characterized in both phyla by expression of either an αβ or a γδ T-cell antigen receptor (TCR). The genes encoding the chicken TCR α- and TCR β-chains, and their mode of repertoire diversification, resemble their mammalian counterparts (2–4). However, the chicken TCR α and β loci are relatively simple in that each contains only two V subfamilies (3–5) versus the 20–30 subfamilies of Vα and Vβ genes found in mice and humans (6, 7). Interestingly, αβ T cells that express the prototypic Vβ1 genes migrate preferentially to the chicken intestine, where they provide help to mucosal B cells for IgA antibody production (8).
Avian T cells bearing a γδ TCR are the first to be generated during ontogeny (9) and they comprise up to 50% of the recirculating T-cell pool in mature birds (9, 10). This relative abundance of γδ versus αβ T cells and the experimental accessibility of avian embryos make the chicken an attractive model in which to explore unresolved issues in γδ T-cell development and function. However, more information on the TCR γδ genes is needed to exploit this avian model. A chicken TCRγ gene candidate has been isolated by PCR using short, minimally degenerate oligonucleotide primers complementing conserved V region segments to amplify TCR-like products from genomic DNA (11, 12). The present studies refine the definition of this candidate TCRγ gene, outline the composition of the chicken TCRγ locus, determine the embryonic pattern of TCRγ gene expression, and examine the initial TCRγ repertoire diversification in the thymus.
MATERIALS AND METHODS
Chickens and Cell Lines.
Inbred SC (Hyline International, Dallas, IA) and H.B19ov+ (Basel Institute for Immunology, Switzerland) White Leghorn chickens were used. The CU15 αβ T cell line was transformed by Marek disease virus (13), the DT40 avian leukosis virus-induced bursal lymphoma B cell line was from E. Humphries (14), and the 857–7 γδ T-cell line, transformed with reticuloendotheliosis virus strain T, was the kind gift of T. Graf and K. McNagny (European Molecular Biology Laboratory, Heidelberg).
cDNA and Genomic Library Screening.
A chicken spleen library, made in the Uni-ZAP XR vector (Stratagene) (5), was screened according to manufacturer’s recommendations, and pBluescript subclones were prepared for sequencing. A chicken liver genomic pWE15 library (CLONTECH) was screened according to the manufacturer’s protocol. Isolated cosmid clones were analyzed by restriction mapping and Southern blotting. Hybridizing fragments were subcloned into pBluescript (Stratagene) for sequencing.
Northern and Southern Blots.
Cellular RNA was isolated by the guanidine thiocyanate/cesium chloride method (15). RNA from embryonic thymus samples were extracted with TRI REAGENT (Molecular Research Center, Cincinnati). DNA was prepared as described (16). Northern and Southern blot hybridizations (15) used Magna nylon membranes (Micron Separations, Westboro, MA) at 42°C in 50% formamide buffer and DNA probes 32P-labeled with a Prime-It II random primer labeling kit (Stratagene). Low stringency washing conditions used 2× SSC and 0.05% SDS at 42°C. For rehybridization, membranes were stripped in 0.05% SDS at 90°C for 15 min.
Reverse Transcription–PCR (RT-PCR) and Anchored-PCR.
cDNA was synthesized from 5–10 μg of total RNA using Superscript reverse transcriptase (GIBCO/BRL). PCR was carried out using GIBCO/BRL Taq DNA polymerase under the following cycling conditions: 1 min at 94°C; 30 to 35 cycles of 1 min at 94°C, 1 min at 55°C, 1 min at 72°C; and 7 min extension at 72°C. For the first round of anchored-PCR (17), poly(G)-tailed cDNA was amplified with XNSC10 and Cγdown2 primers (see below). Purified PCR products were reamplified with the XNSC10 and Cγdown1 primers. The final PCR band was purified, and cloned in SmaI-digested, dephosphorylated pBluescript.
Sequencing and Sequence Analysis.
Double-stranded pBluescript templates were prepared for sequencing by the alkaline lysis method (15). Insert nucleotide sequences were determined using the Sequenase 2.0 kit (United States Biochemical). The dnastar software package (DNAstar, Madison, WI) was used for sequence analysis. Data base searches were performed on the Internet blast server at the National Center for Biotechnology Information (18).
Primers and DNA Probes.
The 5′ to 3′ sequence PCR primers correspond to the coding strand (up) or the complementary strand (down); Cγdown1, GACTCGAGCTCTCCAGTGGTACAGATAAC; Cγdown2, CTGAGCTCGAGGAGACCTCTCTGAAGAAG; Vγ1 up2, CTGCTACCAGAGAGAGATCC; Vγ2 up1, CCAAAGGCACAGATACAGG; Vγ3 up2, AAGAGGATACTGTACATGTC; XNSC10, CACTCGAGCGGCCGCGTCGACCCCCCCCCCC. The Cγ probe was derived by PCR from the Gd186 cDNA clone using Cγup1 (TGCAGGAGGAACATGAA) and Cγdown3 (GCTTAGCTGCAGTCCTTG) primers. Vγ1 and Vγ2 probes corresponded to Gd186 and Gd187 PCR fragments (12). The Vγ3 probe was derived from 9a1 cDNA clone by amplification with Vγ3down1 (GAGTTGGAAGGATTTCTCTGC) and pBluescript RPSK (GGAAACAGCTATGACCATGA) primers. The chicken glyceraldehyde- 3-phosphate dehydrogenase cDNA probe was isolated in our laboratory.
RESULTS
Characterization of a Full-Length TCRγ cDNA.
PCR-derived candidates for the chicken TCR γ-chain, Gd186 and Gd187 (12), share 43% and 25% sequence identity at the nucleic acid and the amino acid levels, respectively, and contain key residues at positions that define TCR and Ig variable domains. Neither candidate is related closely to chicken TCRα, TCRβ, or Ig sequences, suggesting they may code for variable regions of TCR γ- or TCR δ-chains.
DNA probes derived from the Gd186 and Gd187 M13 clones were used to screen a chicken spleen cDNA library. The sequence of one of the largest Gd186-hybridizing cDNA clones consisted of 1920 bp with an open reading frame of 344 amino acids flanked by a 3′ untranslated region of 877 bases (Fig. 1). A consensus polyadenylylation sequence (AATAAA) is found 10 nucleotides upstream of the beginning of the poly(A) tail. The variable domain exhibits 30–35% identity with mammalian Vγ gene segments, similar to that found between variable regions of the avian and mammalian TCRαβ genes (3, 4). The variable region also contains conserved residues characteristic of TCR/Ig variable regions (11, 20), Cys-21, Trp-Tyr-33, Gln-35, Tyr-46, Asp-84, Tyr-Tyr-Cys-90, and of residues characteristic of the TCRγ variable regions, Ser-8, Tyr-Ile-His-31, Lys-61, Trp-93 (6, 7, 21). A J-like region (residues 100 to 116A) shares 70% identity at the amino acid level with mammalian TCR Jγ segments, including the canonical J segments motif, Phe-Gly-Xaa-Gly-Thr-112 (21).
Figure 1.

Nucleotide and predicted amino acid sequence of 1.9-kb TCRγ G186cDNA (GenBank accession no. U22666U22666). Boundaries between the leader peptide (L), variable (V), junctional (J), extracellular (Cex), transmembrane (TM), and cytoplasmic (Cyt) regions are indicated by arrow heads. Boundary and canonical residues are numbered according to mammalian Vγ gene segment convention (6, 7). The 51-residue leader peptide was predicted by the psort program (http://psort.nibb.ac.jp) based on the criteria of von Heijne (19). Putative CDR1 and CDR2 regions are indicated. Asp117 is the first position assigned to the Cγ region, although in TCR γ-chains using Jγ3 the aspartic acid is replaced by an asparagine (Fig. 3B). Residues conserved in mammalian Cγ genes are underlined. One potential N-glycosylation site (★) is found at position 237. A consensus polyadenylylation sequence is boldfaced.
The region corresponding to residues 117 to 289 shares 25–30% identity at the amino acid level with mammalian TCR Cγ regions (12). A predicted transmembrane region (residues 252 to 274) contains a conserved lysine residue thought to interact with negatively charged residues in CD3 molecules. Like mammalian TCR γ-chains, Gd186 cDNA contains a predicted intracellular domain of 15 residues, whereas other Ig/TCR chains have cytoplasmic tails of less than 6 residues. The extracellular portion consists of 135 residues. Two cysteine residues, Cys-141 and Cys-200, believed to form the intrachain disulfide bond of the Cγ domain, are spaced by 59 amino acids as compared with the 55 residue spacing in mammalian Cγ regions (23). An additional cysteine residue at position 231 is predicted to form the interchain disulfide bond with the TCR δ-chain. This cysteine is located within the connecting peptide, predicted by comparison with mammalian sequences to extend from residues 223 to 251 (Fig. 1). Connecting peptides of mammalian Cγ genes exhibit little sequence similarity (23); when this region is omitted from the comparison, the putative chicken Cγ gene shares 25–35% identity with mammalian Cγ genes. Transmembrane and cytoplasmic regions exhibit 30–50% and 20–45% identity, respectively. Amino acid sequence analysis of the chicken Cγ candidate reveals 48 positions that are conserved in 80% of mammalian Cγ genes, 17 being invariant (underlined in Fig. 1).
The predicted molecular mass of the mature polypeptide is 33 kDa, in close agreement with the 34-kDa molecular mass estimate for chicken TCR γ-chains after removal of carbohydrate (9), presumably attached to a potential N-glycosylation site at position 236. The predicted isoelectric point of 7.95 for the Gd186 cDNA mature protein is consistent with the γ-chain being more basic than the δ-chain (24, 25).
Tissue Distribution of Candidate TCRγ Gene Transcripts.
When a probe corresponding to the predicted Cγ region of Gd186 cDNA was used for Northern blot analysis, a 1.9-kb transcript was detected in cells from the thymus, spleen, peripheral blood, cecal tonsils, and a γδ T-cell line. A weak hybridizing band was also detected in liver and bursa, which may contain a few T cells, whereas transcripts were not detected in the DT40 B cell line or the CU15 αβ T-cell line (Fig. 2). Transcripts detected by the candidate Cγ probe thus appear confined to γδ T cells.
Figure 2.
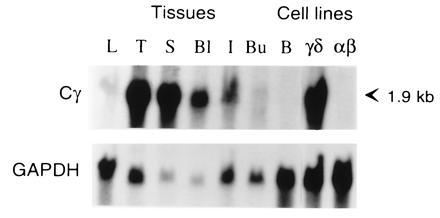
Northern blot analysis of different tissues and cell lines with the candidate TCR Cγ cDNA probe. RNA (10 μg) from liver (L), thymus (T), spleen (S), blood lymphocytes (Bl), intestinal cecal tonsils (I), bursa (Bu), DT40 B cell line (B), 857–7 γδ cell line (γδ), and CU15 αβ cell line (αβ) was electrophoresed in agarose, blotted on nylon membrane, and sequentially hybridized with 32P-labeled Cγ (G186cDNA origin) and glyceraldehyde-3-phosphate dehydrogenase probes.
Identification of Three Vγ Subfamilies and Three Jγ Gene Segments.
Vγ subfamilies related to the Gd186 and Gd187 PCR products were designated Vγ1 and Vγ2, respectively. Additional cDNA clones were identified in the chicken spleen library by differential screening with the Cγ, Vγ1, and Vγ2 probes using low stringency washing conditions. This allowed cross-hybridization with genes sharing at least 75% identity, a value delineating members within the same subfamily (26). Additional Vγ1+ and Vγ2+ clones were identified in this way. Two Vγ1− Vγ2− Cγ1+ clones, 5d1 and 9a1, contained variable gene segments defining a third Vγ subfamily designated Vγ3 (Fig. 3A). In addition to the Jγ region identified in clone Gd186 cDNA, designated Jγ1, two other Jγ segments, Jγ2 and Jγ3, were revealed by analysis of cDNA clones (Fig. 3B). Examination of the CDR3 regions of these clones suggests exonuclease activity, possible P-addition and N-addition (Fig. 3B), in keeping with the principles of V(D)J gene segment rearrangement in other TCR loci (27).
Figure 3.
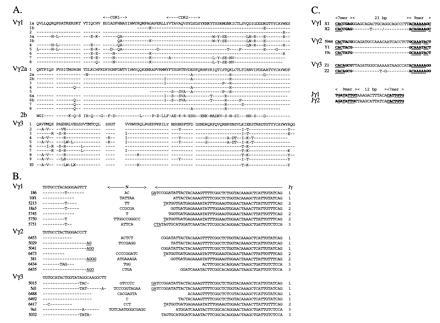
TCR Vγ and Jγ nucleotide and amino acid sequences. (A) Comparison of amino acid sequences of Vγ1, Vγ2, and Vγ3 subfamily members characterized according to sequence data of genomic gene segments, cDNA clones from the cDNA library, and from anchored-PCR or specific PCR. Residues identical to the prototypic sequence are indicated by dashes. Vγ1.1b, Vγ1.3b, and Vγ2.5 are partial amino sequences. (B) Nucleotide sequences of Vγ–Jγ junctions. Vγ usage is indicated on the left and Jγ usage on the right. Vγ, Jγ1, and Jγ2 gene segment assignment is based on germ-line sequences. Dashes indicate identity to the germ-line Vγ sequence. Assignment of nucleotides to the Jγ3 gene segment is based on a consensus sequence of TCR γ-chains using the Jγ3 segment. Nucleotides that cannot be assigned to either Vγ or Jγ gene segments are indicated as N- or P-additions. Putative P nucleotides are underlined. (C) Recombination signal sequences of Vγ1, Vγ2, Vγ3, Jγ1, and Jγ2 gene segments. Heptamer/nonamer sequences, indicated in boldface type, were identified by comparison to the consensus sequences of mammalian TCR/Ig genes (27). Positions matching the consensus sequences are underlined.
Germ-Line Sequences of Chicken TCR Vγ and Jγ Gene Segments.
Genomic Vγ1, Vγ2, and Vγ3 gene segments were identified by screening a liver genomic cosmid library with Vγ probes. Recombination signal sequences, including 23-bp spacer sequence, are conserved for each Vγ subfamily, but differ between subfamilies (Fig. 3C). Recombination signal sequences, including 12-bp spacer sequences, were also identified for Jγ1 and Jγ2 segments by sequencing a ≈3.3-kb PCR fragment obtained after amplification of chicken genomic DNA (Fig. 3C). None of the recombination signal sequences match the consensus sequences precisely, but the positions required for efficient recombination are present (27).
The chicken TCRβ, and mammalian TCR variable gene segments exhibit a typical two exon structure spaced by approximately 100 bp of intronic sequence, whereas the chicken Vα1 genes are encoded by a single exon (4). In this context, chicken Vγ3 gene segments display the two exon structure typical of mammalian TCR V genes, while both the Vγ1 and Vγ2 genomic gene segments were found to consist of a single exon (GenBank accession nos. U78251–U78256, U78251, U78252, U78253, U78254, U78255, U78256).
Genomic and cDNA Analysis Indicates a Single Cγ Gene and Multiple Members of the Three Vγ Subfamilies.
When genomic DNA was analyzed by Southern blotting to search for Cγ genes, a single hybridization band was observed with PstI (5.6 kb) and XbaI (5.9 kb) digests, even under low stringency conditions (Fig. 4, and data not shown). Two or three smaller fragments were detected with EcoRI (3.3 and 2.4 kb) and HindIII (1.7, 1.3, and 0.6 kb). Since mammalian Cγ genes and the chicken Cα and Cβ genes span several kilobases of genomic DNA (ref. 28; unpublished observations), these data suggest that the chicken TCRγ locus contains only one Cγ gene.
Figure 4.
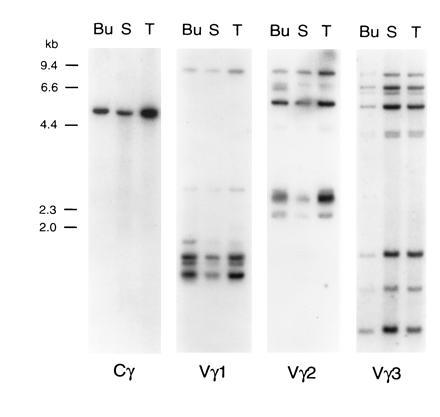
Southern blot analysis of chicken genomic DNA with TCRγ constant and variable region probes. DNA (15 μg) from bursa (Bu), spleen (S), and thymus (T) was digested with the PstI restriction enzyme, electrophoresed in agarose, and blotted onto nylon membranes. Hybridization was performed with the Cγ probe and, after stripping, with Vγ1, Vγ2, and Vγ3 probes. Sizes of λ HindIII DNA marker fragments are indicated on the left. Digestion with EcoRI, XbaI, and HindIII restriction enzymes gave similar results (data not shown).
Genomic DNA digested with four restriction enzymes and hybridized with prototypic Vγ1, Vγ2, and Vγ3 probes reveals 7–9 hybridizing bands (Fig. 4, and data not shown). The relatively high intensity of some hybridizing bands suggests the possible superimposition of DNA fragments of similar length. Similarities between the hybridization patterns obtained with the different Vγ probes (e.g., an 8.1-kb PstI fragment noted with both the Vγ1 and Vγ2 probes and fragments of 7.4, 7.2, 6.4, and 5.2 kb with the Vγ2 and Vγ3 probes) suggest interspersion of members of different Vγ subfamilies. This suggestion was confirmed by demonstration that individual genomic cosmid clones of ≈40 kb hybridized with the Vγ1, Vγ2 and Vγ3 probes, and that the same 4-kb EcoRI–XbaI band contained members of both the Vγ1 and Vγ3 subfamilies (data not shown).
Analysis of other cDNA clones from embryonic thymus, adult spleen and adult intestine, obtained by specific PCR using Vγ1, Vγ2, Vγ3, and Cγ primers, or anchored-PCR, indicate 8–10 members in each Vγ subfamily (Fig. 3A). In this analysis, sequences differing by more than one amino acid were considered diagnostic of different genes. Chicken Vγ1 members share 79% and 91% identity at the amino acid and nucleotide levels, respectively. TCRγ transcripts cloned by anchored PCR from the 857–7 γδ cell line were found to contain a variable gene segment related to the Vγ2 subfamily (clone 383). All Vγ2 members, except clone 383, share at least 85% and 92% identity at the amino acid and nucleotide levels. Clone 383 shares 56–59% residues and 77% nucleotide sequence identity with other Vγ2 members. The other Vγ2 clones were therefore designated Vγ2a subfamily members and clone 383 a Vγ2b subfamily member. Vγ3 members share more than 82% and 91% identity at the amino acid and nucleic acid levels, respectively. The degree of sequence similarity between the three Vγ subfamilies is similar to that seen between subfamilies of other chicken and mammalian TCR variable gene segments. Conserved residues are limited largely to canonical residues defined for TCR/Ig sequences.
All Three Vγ Subfamilies Participate in Embryonic Repertoire Diversification.
When transcriptional activity of rearranged Vγ1, Vγ2, and Vγ3 genes was assessed by RT-PCR in the embryonic thymus, transcripts were detected for each of the Vγ subfamilies beginning on embryonic day 10 (E10) indicative of parallel usage of the Vγ subfamilies during intrathymic ontogeny (Fig. 5). TCR Vβ1 transcripts were detected by E12–E13, whereas the onset of TCR Vβ2 transcription was not evident until E14, in keeping with earlier analyses of TCR Vβ1 and Vβ2 rearrangement and expression (24, 29–31). To examine further the pattern of TCRγ repertoire diversification during the first wave of thymocyte ontogeny, Vγ1-Cγ, Vγ2-Cγ, and Vγ3-Cγ PCR products from the E10 embryos were cloned and sequenced. Analysis of the E10 transcripts (GenBank accession nos. U78210–U78230, U78210, U78211, U78212, U78213, U78214, U78215, U78216, U78217, U78218, U78219, U78220, U78221, U78222, U78223, U78224, U78225, U78226, U78227, U78228, U78229, U78230) confirmed the non-preferential usage of Vγ subfamily members. Examination of the CDR3 regions indicates the presence of P-nucleotides or variable nuclease activity, and limited N-region addition compared with the adult repertoire. These observations indicate the generation of a surprisingly diverse TCRγ repertoire during the initiation of the first embryonic thymocyte wave.
Figure 5.
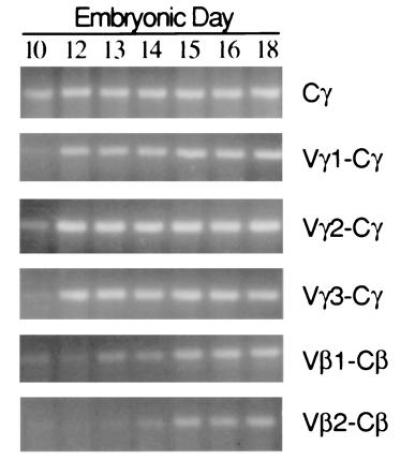
Ontogenetic pattern of Vγ and Vβ expression in the thymus. Thymi were collected from individual embryos at 10 to 18 days of incubation. RNA was extracted from homogenized tissues and first-strand cDNA was synthesized. PCR assays were conducted with relevant primers to detect TCR Cγ, Vγ1-Cγ, Vγ2-Cγ, Vγ3-Cγ, Vβ1-Cβ, and Vβ2-Cβ transcripts. PCR products were detected after agarose gel electrophoresis and staining with ethidium bromide.
DISCUSSION
A complex organization of the TCRγ gene locus in a representative avian species, G. gallus domesticus, is revealed in this study. The locus consists of three Vγ subfamilies, each of which includes approximately 8–10 members, three Jγ gene segments, and a single Cγ gene. The Vγ subfamily members recombine with each of the Jγ gene segments early in ontogeny to generate a highly diverse repertoire of TCR γ-chains.
Conservation of the Structural Features of TCR Chains.
Our characterization of the chicken TCRγ locus confirms the emergence of αβ and γδ T cell sublineages before the divergence of birds and mammals more than 200 million years ago. A salient feature indicated by this analysis is the striking conservation of the overall structure of the avian TCRγ genes, despite the limited number of residues shared with mammalian TCRγ genes. Most conserved residues are not specific for TCR γ-chains, but rather are a general characteristic of Ig/TCR chains. These include the conserved cysteines in the variable and constant domains involved in the intrachain and interchain disulfide bonds and the FGXGT motif characteristic of J gene segments. This pattern of conservation, reminiscent of that seen for the TCRα and TCRβ genes in fish, amphibians, birds, and mammals (3, 4, 11, 32–34), makes it difficult to identify the Ig/TCR family solely by sequence comparison. Multiple alignment of sequences nevertheless revealed motifs characteristic of Ig/TCR families that helped to implicate cDNA Gd186 as a TCRγ family member. In particular, the CART motif (Conserved Antigen Receptor Transmembrane) characteristic of TCRγ/β-chains can be detected in the chicken Cγ gene. The CART motif has been found in the transmembrane domains of all antigen receptors thus far identified in vertebrates and is considered essential for receptor assembly and/or signaling (35).
Comparison of the chicken Jγ gene segments with known mammalian Jγ segments indicates a recurrent consensus sequence that includes the FGXGCT motif in the framework 4 region of other TCR or Ig chains. Chicken Jγ segments have complete identity with the consensus sequence. Notably, the TCR Jγ-characteristic lysine residue at position 106, conserved in all but one Jγ segment, is never found in other TCR/Ig J-gene segments. Analysis of the chicken TCRα and TCRβ loci also indicates a high conservation of Jα and Jβ gene segments (3, 4). The three chicken Jγ segments share 57–63% identity at the nucleotide sequence level and 63–75% identity at the amino acid level, and are more closely related to each other than to any of the Jγ segments in mammalian species. This structural conservation is particularly evident in the framework 4 region, where most substitutions are silent. These observations suggest the triplication of a single Jγ ancestral gene in the chicken, but would also be consistent with selective pressure acting to conserve the framework region of J gene segments.
Organization of the Chicken TCR Locus.
While TCRγ genes have been characterized in several mammalian species, the genomic organization of the TCRγ loci is known only for mice and humans (28, 36). In humans, eight functional Vγ gene segments can recombine with five Jγ segments organized in two clusters, each associated with one Cγ gene (36) to yield 40 potential V-J pairings. In mice, seven Vγ and four Jγ gene segments are organized in four clusters of V-J-Cγ genes (36, 37). Rearrangements between gene segments belonging to different clusters are infrequent, so that only seven V-Jγ combinations are ordinarily used in murine TCR γ-chains. Sequence analysis of sheep and cow cDNA also suggests that the Vγ and Jγ gene segments are organized in recombination clusters so that particular Vγ subfamilies are always found rearranged to the same Jγ-Cγ genes (23). Our analysis indicates that the chicken has three multimember Vγ gene subfamilies, three Jγ gene segments, and one Cγ gene. Genomic DNA analysis indicates that the three Jγ gene segments are located within ≈3.3 kb. Each Jγ segment recombines with members of all three Vγ subfamilies, indicating that the chicken Vγ and Jγ gene segments are not organized in clusters that preclude segmental rearrangement. The chicken TCRγ locus thus resembles the human TCRγ locus in this respect. Unlike the mouse and human TCRγ loci, however, all three chicken Vγ subfamilies appear to contain a relatively large number of members, approximately 8 to 10, that may be interspersed with members of other subfamilies. This predicts a genomic organization in which clusters containing members of each Vγ subfamilies are repeated several times, as is the case for TCR Vα and Vβ subfamilies in mouse and humans (38, 39), and implies successive duplications of ancestral Vγ1, Vγ2, and Vγ3 gene segments.
TCRγ Repertoire Diversification Early in Avian Ontogeny.
A highly regulated pattern of rearrangement and expression of the different Vγ genes during murine ontogeny is reflected by waves of tissue-specific migration by γδ T cells with canonical Vγ-Jγ rearrangements (36). The Vγ3-Jγ1-Cγ1-bearing T cells home to the skin epithelium, whereas γδ T cells with Vγ5-Jγ1-Cγ1 rearrangements are seeded preferentially to the intestine. Likewise, Vβ1 subfamily members in the chicken are programmed for intrathymic rearrangement before the Vβ2 subfamily members (31), and the Vβ2+ αβ T cells rarely migrate to the intestine (8). In contrast, analysis of the TCRγ transcripts in chicken embryos indicates that rearrangement of all three Vγ subfamilies begins in the day 10 embryonic thymus without preference for a particular Vγ subfamily. This early diversification of the V-Jγ junctions also involves variable exonuclease activity, P-region addition, and even a limited degree of N-region addition. The TCRγ repertoire of the first wave of thymocytes in the chicken is thus remarkably diverse and unrestricted in comparison with the mouse.
Relationship Between the Complexity of the TCRγ Loci and the Relative Abundance of γδ T Cells.
The high frequency of γδ T cells in chickens, cattle, sheep, and pigs (9, 23, 40, 41) is associated with more complex TCRγ and TCRδ loci in these species than in mice and humans. Conversely, there are fewer Vβ, Vα, Jβ, and Jα gene segments in chickens than in mice and humans (1). The numbers of possible TCR αβ and γδ V(D)J rearrangements thus may correlate with the relative frequencies of the αβ and γδ T cells. The Ig and TCRαβ loci appear to have emerged concomitantly during the evolution of the first jawed vertebrates (42, 43). A series of gene expansion–contraction events coupled to selection (44) would thus be expected to result in variable degrees of TCR/Ig complexity in extant representatives of the different vertebrate phyla. In accordance with this theory, relatively simple TCRαβ loci characterize the chicken, whereas extensive genetic diversity has been described in shark (45), trout (32, 34), axolotl (33), and mammals (6, 7). Similarly, the TCRγ locus is relatively complex in chickens, cattle, sheep, and pigs in comparison with mice and humans. Definition of this locus in other vertebrate representatives may lead to better understanding of the γδ T cells and their functional relevance.
Acknowledgments
We thank Yolanda Hartman, Jinyi Wang, and Colette Jacono for technical assistance, Robin Dzialo for advice and help, Ann Brookshire for secretarial assistance, and Drs. Peter D. Burrows, Richard D. Hockett, Vincent Hurez, Gary W. Litman, and Harry W. Schroeder, Jr., for reading the manuscript. This work was supported in part by the National Institutes of Health Grants AI34008 (W.T.M.) and AI30879 (M.D.C.), by the Association pour la Recherche contre le Cancer, and by the Human Frontier Science Program Organization. A.S. is a Research Associate, and M.D.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Abbreviations: TCR, T cell antigen receptor; RT-PCR, reverse transcription–PCR.
Data deposition: The sequences reported in this paper have been deposited in the GenBank data base (accession nos. U78207–U78288, U78207, U78208, U78209, U78210, U78211, U78212, U78213, U78214, U78215, U78216, U78217, U78218, U78219, U78220, U78221, U78222, U78223, U78224, U78225, U78226, U78227, U78228, U78229, U78230, U78231, U78232, U78233, U78234, U78235, U78236, U78237, U78238, U78239, U78240, U78241, U78242, U78243, U78244, U78245, U78246, U78247, U78248, U78249, U78250, U78251, U78252, U78253, U78254, U78255, U78256, U78257, U78258, U78259, U78260, U78261, U78262, U78263, U78264, U78265, U78266, U78267, U78268, U78269, U78270, U78271, U78272, U78273, U78274, U78275, U78276, U78277, U78278, U78279, U78280, U78281, U78282, U78283, U78284, U78285, U78286, U78287, U78288).
References
- 1.Chen C H, Six A, Kubota T, Tsuji S, Kong F, Göbel T W F, Cooper M D. Curr Top Microbiol Immunol. 1996;212:37–53. doi: 10.1007/978-3-642-80057-3_5. [DOI] [PubMed] [Google Scholar]
- 2.Cooper M D, Chen C H, Bucy R P, Thompson C B. Adv Immunol. 1991;50:87–117. doi: 10.1016/s0065-2776(08)60823-8. [DOI] [PubMed] [Google Scholar]
- 3.Tjoelker L W, Carlson L M, Lee K, Lahti J, McCormack W T, Leiden J M, Chen C H, Cooper M D, Thompson C B. Proc Natl Acad Sci USA. 1990;87:7856–7860. doi: 10.1073/pnas.87.20.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Göbel T W F, Chen C H, Lahti J, Kubota T, Kuo C-L, Aebersold R, Hood L, Cooper M D. Proc Natl Acad Sci USA. 1994;91:1094–1098. doi: 10.1073/pnas.91.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubota T, Chen C H, Hockett R, Göbel T W F, Cooper M D. FASEB J. 1995;9:A817. (abstr. 4736). [Google Scholar]
- 6.Arden B, Clark S P, Kabelitz D, Mak T W. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 7.Arden B, Clark S P, Kabelitz D, Mak T W. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 8.Cihak J, Hoffmann-Fezer G, Ziegler-Heitbrock H W L, Stein H, Kaspers B, Chen C H, Cooper M D, Lösch U. Proc Natl Acad Sci USA. 1991;88:10951–10955. doi: 10.1073/pnas.88.23.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sowder J T, Chen C H, Ager L L, Chan M M, Cooper M D. J Exp Med. 1988;167:315–322. doi: 10.1084/jem.167.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cihak J, Lösch U, Hoffmann-Fezer G, Chen C H, Cooper M D, Ziegler-Heitbrock H W L. Scand J Immunol. 1993;38:123–129. doi: 10.1111/j.1365-3083.1993.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 11.Rast J P, Litman G W. Proc Natl Acad Sci USA. 1994;91:9248–9252. doi: 10.1073/pnas.91.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rast J P, Haire R N, Litman R T, Pross S, Litman G W. Immunogenetics. 1995;42:204–212. doi: 10.1007/BF00191226. [DOI] [PubMed] [Google Scholar]
- 13.Schat K A, Chen C H, Calnek B W, Char D. J Virol. 1991;65:1408–1413. doi: 10.1128/jvi.65.3.1408-1413.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baba T W, Humphries E H. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 15.Ausubel F M, Brent R, Kingston R E, Moore D E, Smith J A, Seidman F G, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 16.Thompson C B, Neiman P E. Cell. 1987;48:369–378. doi: 10.1016/0092-8674(87)90188-7. [DOI] [PubMed] [Google Scholar]
- 17.Six A, Jouvin-Marche E, Loh D Y, Cazenave P-A, Marche P N. J Exp Med. 1991;174:1263–1266. doi: 10.1084/jem.174.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chothia C, Boswell D R, Lesk A M. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of Proteins of Immunological Interest. Natl. Inst. Health, Bethesda: Public Health Service; 1991. [Google Scholar]
- 22.Frank S J, Engel I, Rutledge T M, Letourneur F. Semin Immunol. 1991;3:299–311. [PubMed] [Google Scholar]
- 23.Hein W R. Semin Immunol. 1994;6:361–372. doi: 10.1006/smim.1994.1046. [DOI] [PubMed] [Google Scholar]
- 24.Char D, Sanchez P, Chen C H, Bucy R P, Cooper M D. J Immunol. 1990;145:3547–3555. [PubMed] [Google Scholar]
- 25.Moretta L, Ciccone E, Ferrini S, Pelicci P G, Mingari M C, Zeromski J, Bottino C, Grossi C, Moretta A. Immunol Rev. 1991;120:117–135. doi: 10.1111/j.1600-065x.1991.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 26.Brodeur P H, Riblet R. Eur J Immunol. 1984;14:922–930. doi: 10.1002/eji.1830141012. [DOI] [PubMed] [Google Scholar]
- 27.Lewis S M. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 28.Vernooij B T M, Lenstra J A, Wang K, Hood L. Genomics. 1993;17:566–574. doi: 10.1006/geno.1993.1373. [DOI] [PubMed] [Google Scholar]
- 29.Lahti J M, Chen C H, Tjoelker L W, Pickel J M, Schat K A, Calnek B W, Thompson C B, Cooper M D. Proc Natl Acad Sci USA. 1991;88:10956–10960. doi: 10.1073/pnas.88.23.10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunon D, Schwager J, Dangy J-P, Imhof B A. J Immunol. 1995;154:1256–1264. [PubMed] [Google Scholar]
- 31.Bucy R P, Chen C H, Cooper M D. J Immunol. 1990;144:1161–1168. [PubMed] [Google Scholar]
- 32.Partula S, De Guerra A, Fellah J S, Charlemagne J. J Immunol. 1995;155:699–706. [PubMed] [Google Scholar]
- 33.Fellah J S, Kerfourn F, Charlemagne J. J Immunol. 1994;153:4539–4545. [PubMed] [Google Scholar]
- 34.Partula S, De Guerra A, Fellah J S, Charlemagne J. J Immunol. 1996;157:207–212. [PubMed] [Google Scholar]
- 35.Campbell K S, Bäckström B T, Tiefenthaler G, Palmer E. Semin Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- 36.Raulet D H. Annu Rev Immunol. 1989;7:175–207. doi: 10.1146/annurev.iy.07.040189.001135. [DOI] [PubMed] [Google Scholar]
- 37.Pereira P, Gerber D, Regnault A, Huang S Y, Hermitte V, Coutinho A, Tonegawa S. Int Immunol. 1996;8:83–90. doi: 10.1093/intimm/8.1.83. [DOI] [PubMed] [Google Scholar]
- 38.Jouvin-Marche E, Hue I, Marche P N, Liebe-Gris C, Marolleau J-P, Malissen B, Cazenave P-A, Malissen M. EMBO J. 1990;9:2141–2150. doi: 10.1002/j.1460-2075.1990.tb07383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rowen L, Koop B F, Hood L. Science. 1996;272:1755–1762. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- 40.Thome M, Hirt W, Pfaff E, Reddehase M J, Saalmüller A. Vet Immunol Immunopathol. 1994;43:13–18. doi: 10.1016/0165-2427(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 41.Hein W R, Mackay C R. Immunol Today. 1991;12:30–34. doi: 10.1016/0167-5699(91)90109-7. [DOI] [PubMed] [Google Scholar]
- 42.Marchalonis J J, Schluter S F. Scand J Immunol. 1990;32:13–20. doi: 10.1111/j.1365-3083.1990.tb02886.x. [DOI] [PubMed] [Google Scholar]
- 43.Thompson C B. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 44.Ota T, Nei M. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- 45.Hawke N A, Rast J P, Litman G W. J Immunol. 1996;156:2458–2464. [PubMed] [Google Scholar]


