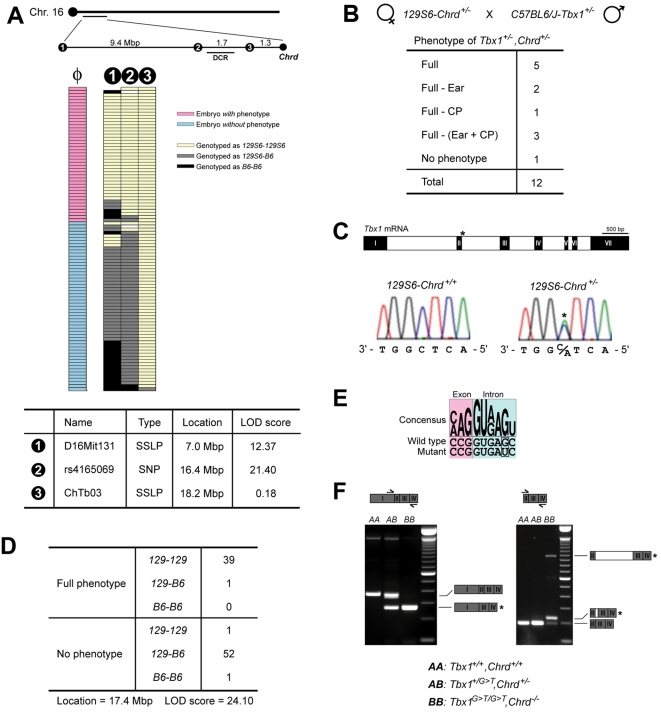
Figure 2. Characterization of a linked modifier of Chrd in the 129S6 strain.
(A) Schematic diagram of mouse chromosome 16, showing three genetic markers used and genotyping results of F2 hybrid DNAs using these markers. Information on the markers is shown in the table at the bottom of the panel. The second marker rs4165069 shows the strongest co-segregation with the trait. The novel SSLP marker ‘ChTb03’ produces a 271 bp long PCR fragment from 129S6, and 262 bp from B6 (Figure S4). (B) Cross between 129S6-Chrd+/− and B6-Tbx1+/− mice produced Chrd+/−,Tbx1+/− embryos with highly penetrant 22q11DS phenotypes, supporting the existence of modifier linked to Chrd and suggesting the possibility of a mutation in Tbx1 itself. (C) Sequencing of theTbx1 locus of 129S6-Chrd+/+ and 129S6-Chrd+/− strains, revealing a specific point mutation (Tbx1G>T) located in the second intron of the 129S6-Chrd+/− allele (asterisk). (D) Genotyping results of F2 hybrid DNAs using theTbx1G>T mutation as a SNP marker, showing that it is more strongly linked to the phenotype than rs4165069. (E) Tbx1G>T is located at an exon-intron boundary. Consensus, wild-type, and mutant sequences encompassing the mutation are displayed. (F) RT-PCR analysis demonstrates that Tbx1G>T disrupts normal splicing of Tbx1 in 129S6 mice carrying the Chrd null allele. As a result of the point mutation, both exon skipping (left) and intron retention (right) occur in the generation of Tbx1 mRNA, but very little normal message is produced. Diagrams of mRNA with asterisks (*) denote mutant splicing variants that would invariably produce truncated Tbx1 protein.