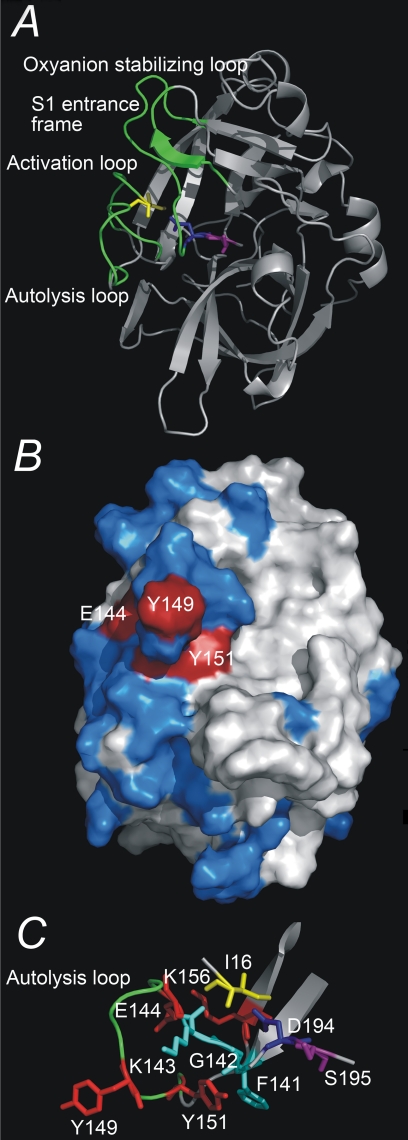
FIGURE 1.
Three-dimensional structure of uPA. A, overview of the three-dimensional structure of the serine protease domain of active uPA, displayed as ribbons. Depicted as sticks are the residues Ile16, Asp194, and Ser195. The activation domain, i.e. the activation loop (residues 16-21), the autolysis loop (residues 142-152), the oxyanion stabilizing loop (residues 184-193), and the S1 entrance frame (residues 216-223) are colored green. B, the epitope of mAb-112, displayed on a surface presentation of the serine protease domain of active uPA. Alanine substitution of residues depicted in red resulted in a significant change in the affinity to mAb-112, whereas alanine substitution of residues depicted in blue did not. C, a close up view of the autolysis loop (residues Gly141 to Lys156) and residues implicated in the binding of mAb-112. All figures were constructed with Pymol on the basis of the coordinates given in the PDB entry 1C5W.