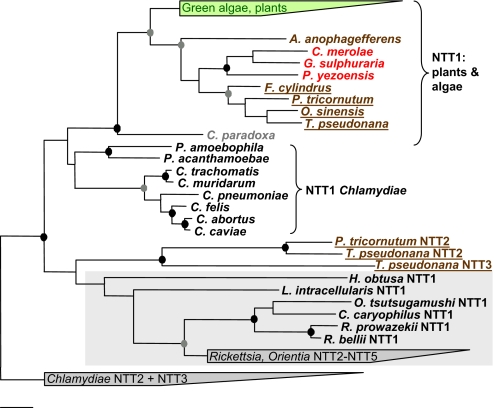
Fig. 4.
Phylogenetic relationships of diatom NTTs with known bacterial and plastidial NTTs. An amino acid-based phylogenetic tree calculated by using the TREE-PUZZLE algorithm is shown. Black circles indicate nodes, which are supported by TREE-PUZZLE support, maximum parsimony, and ProML bootstrap values >80% (1,000 resamplings). Gray circles indicate nodes that are supported by 2 of the 3 above-mentioned treeing methods with support values >80%. TREE-PUZZLE support and bootstrap values <80% are not shown. Bar indicates 10% estimated evolutionary distance. Green algae and plant NTTs are marked in green, red algae are in red, heterokonts are in brown, diatoms are underlined, and C. paradoxa is in light gray. The rickettsial NTT group including NTT1 from L. intracellularis is highlighted by a gray box. A more detailed version of this figure and protein IDs are given in Fig. S6.