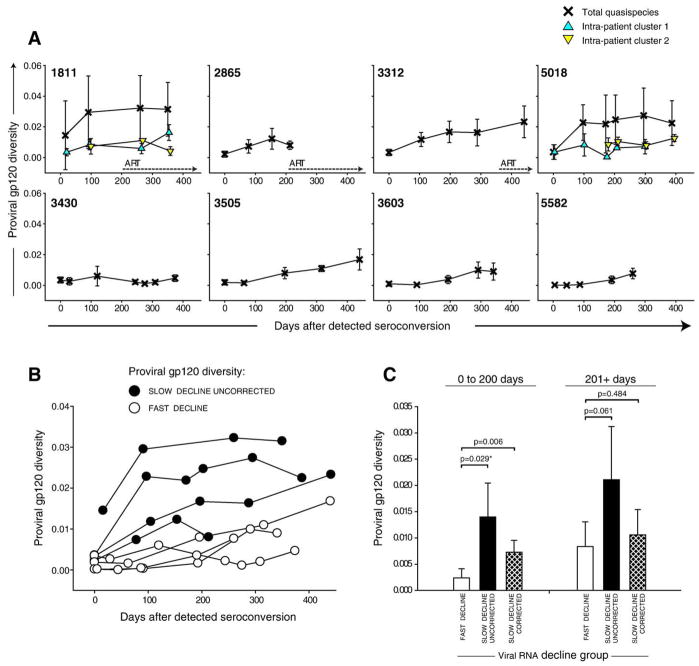
Fig. 5.
Intra-patient viral diversity in gp120. Mean values±standard deviation of pairwise maximum likelihood nucleotide distances are shown. The timeline shows days from detected seroconversion. (A) Individual trajectories of viral diversity in gp120. Numbers within boxes correspond to subject cases. The extent of total viral diversity per time point per subject is shown by crosses. The intra-cluster diversity in subjects 1811 and 5018 is designated by triangles (recombinant sequences are removed). Dotted lines with arrows indicate initiation of ART. (B) Levels of proviral diversity in gp120 between subjects with slow (filled circles) and fast (open circles) decline of viral RNA. (C) Viral diversity within groups defined on viral RNA decline, fast and slow decline of viral RNA. The slow decline group is presented by uncorrected distances (defined as total intra-patient pairwise distances) and cluster-corrected distances (sequences within clusters used separately, no recombinants included). Analysis was performed for two time intervals: 0 to 200 days and 201+ days after seroconversion. Comparison between groups is performed by t-test or Mann–Whitney Rank Sum Test (shown with *).