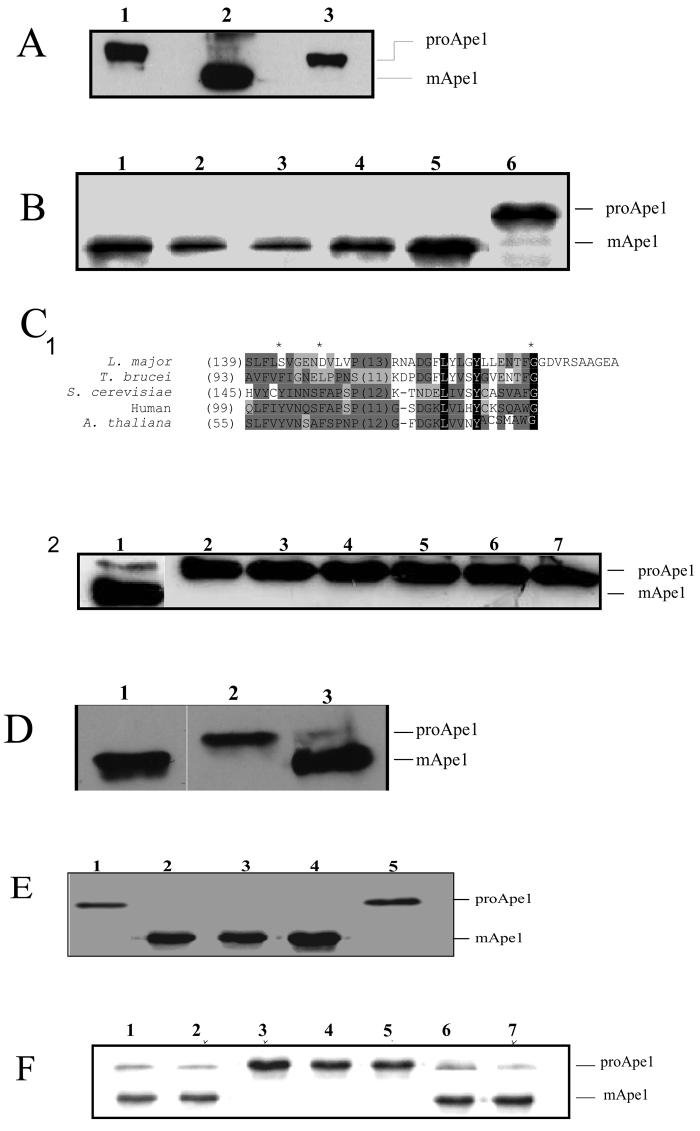
Figure 2. Complementation studies with L. major ATG genes in autophagy-defective Saccharomyces cerevisiae.
A. Western blot analysis of aminopeptidase I (Ape1) in yeast mutant atg8Δ (lane 1), wild type (lane 2) and yeast mutant atg12Δ (lane 3) S. cerevisiae. Cells were incubated under starvation conditions in SD(-N) medium for 16 h, lysed, and API processing was analyzed using rabbit α-Ape1 antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
B. Western blot analysis of Ape1 in atg8Δ yeast transformed with pCM185 containing L. major ATG8 homologues: ATG8 (lane 3), ATG8A (lane 4), ATG8B (lane 5), ATG8C (lane 2) or transformed with pCM185 only (lane 6). Wild type S. cerevisiae are shown in lane 1. The cells were cultured under starvation conditions in SD(-N) medium for 4 h and Ape1 processing was analyzed using rabbit α-API antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
C1. Amino acid sequence alignment of the C-terminal end of L. major ATG12 with those of T. brucei, S. cerevisiae, human and A. thaliana. Identical and conserved amino acids are shaded in black and grey, respectively. Shade of grey depends upon number of residues conserved. The carboxyl-terminal glycine identified in A. thaliana ATG12 (Hanada et al., 2005) and the two hydrophobic residues (Y149 and F154) required for conjugation to ATG10 (Suzuki et al., 2006; Hanada et al., 2006) and essential for the formation of the ATG12-ATG5 conjugate are marked with an asterisk. L. major, LmjF22.1300; T. brucei, Tb927.7.3320; S. cerevisiae, P38316; human, AAH11033; A. thaliana, AAM70187. The numbers in parenthesis represent the amino acid number of the first residue in the alignment.
C2. Western blot analysis of Ape1 in atg12Δ S. cerevisiae transformed with pCM185 containing L. major ATG8 homologues: ATG8 (lane 3), ATG8A (lane 4), ATG8B (lane 5), ATG8C (lane 6), the putative L. major ATG12 (lane 7) or transformed with pCM185 only (lane 2). Wild type S. cerevisiae were used as the control (lane 1). The S. cerevisiae were cultured under starvation condition for 16 h and Ape1 processing was analyzed using a rabbit α-API antibodies. ProApe1 and mature Ape1 (mApe1) are indicated.
D. Western blot analysis of Ape1 in atg5Δ S. cerevisiae transformed with pCM185 containing L. major ATG5 (lane 3) and pCM185 only (lane 2) and treated as described in C2. Wild type S. cerevisiae was included as a positive control (lane 1).
E. Western blot analysis of Ape1 in atg10Δ S. cerevisiae transformed with pCM185 containing the L. major ATG10 homologue (lanes 2-3), or transformed with pCM185 only (lane 1) or ATG8 (lane 5) and treated as described in C2. Wild type S. cerevisiae was included as positive control (lane 4).
F. Western blot analysis of ApeI in atg8Δ S. cerevisiae (lanes 2-4) and atg12Δ S. cerevisiae (lanes 5-7) transformed with pCM185 containing ATG8g (lanes 2, 5), ATG12g (lanes 3, 6) and S. cerevisiae ATG12 (lanes 4, 7), respectively, and treated as described in C2. Wild type S. cerevisiae was included as positive control (lane 1).