Abstract
We present the first case of severe pneumonia possibly caused by Leptotrichia species with oral bacteria. This was found in a healthy but elderly subject whose bronchoalveolar lavage fluid was analyzed by 16S rRNA gene sequencing analysis. The combination of this method and microscopic observation provided useful information for diagnosis and treatment.
CASE REPORT
A 73-year-old Japanese male was admitted to our university hospital for the evaluation and treatment of severe pneumonia with hypoxemia. He had no relevant past medical histories such as periodontitis. He had been in good health until 2 weeks before his first visit to his prior physician, when he developed a sore throat, low-grade fever, nonproductive cough, and serious dyspnea. He initially received cefcapene pivoxil followed by levofloxacin, but his condition deteriorated.
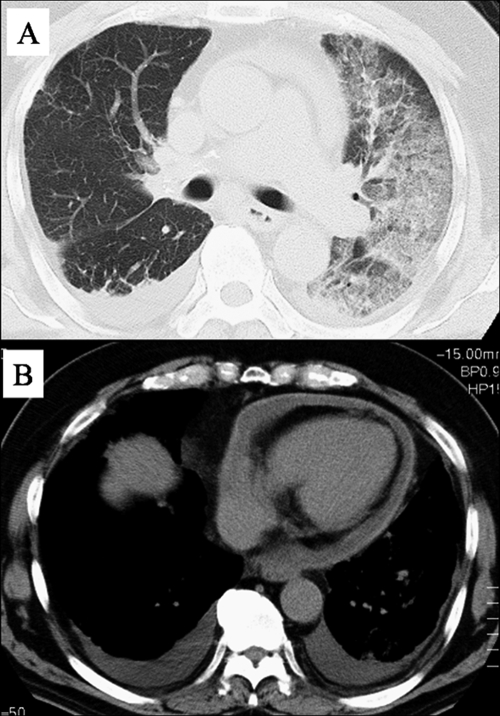
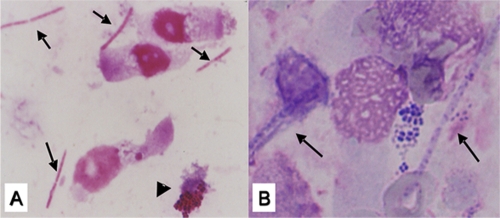
Upon admission to our hospital, inspiratory coarse crackles were audible from the left anterior and posterior thorax. No dental caries and no decayed teeth were seen. His chest X ray and computed tomography showed consolidations surrounded by ground-glass opacities, thickening of interlobular septa in the left lung (Fig. 1A), bilateral pleural effusions, and a pericardial effusion with thickened pericardium (Fig. 1B). Laboratory data showed mild leukocytosis (8,600 white blood cells/μl) without atypical cells including blast cells and elevated C-reactive protein (CRP) (19.4 mg/dl). The partial pressure of arterial oxygen was 64.0 Torr while breathing 4 liters of oxygen per minute by a nasal cannula. Tests for antibodies to Chlamydia pneumoniae, Mycoplasma pneumoniae, human immunodeficiency virus, and human T-cell lymphotropic virus type 1 were negative. Urinary antigens of Legionella pneumophila (BinaxNOW Legionella antigen immunochromatographic test; Binax Inc.) and Streptococcus pneumoniae (BinaxNOW streptococcal antigen immunochromatographic test; Binax Inc.) were not detected. On the day of admission, bronchoalveolar lavage fluid (BALF) was obtained from the left S5 by using fiberoptic bronchoscopy. The recovered fluid contained many neutrophils with numerous gram-negative long rods (approximately 10 μm in length) and some gram-negative and -positive cocci (Fig. 2A). Giemsa staining revealed that the gram-negative long rods had many granules along the long axis (Fig. 2B). However, aerobic cultivation revealed only Enterococcus faecalis. In order to identify the gram-negative rods, the bacterial composition in his BALF was analyzed using a method for clone library sequencing of the 16S rRNA gene.
FIG. 1.
Computed tomography scan of the chest of the subject on admission day illustrating consolidations surrounded by ground-glass opacities and thickening of interlobular septa in the left lung (A) and revealing bilateral pleural effusions and a pericardial effusion with thickened pericardium (B).
FIG. 2.
(A) Gram staining of the BALF obtained from the left S5 using fiberoptic bronchoscopy on admission day revealing gram-negative long rods (arrows) and gram-negative and gram-positive cocci (arrowheads). (B) Giemsa staining of the same specimen showing the long rods with granules along its long axis (arrows).
A 400-μl aliquot of BALF was suspended in 500 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA-2Na [pH 8.0]), 100 μl of 30% sodium dodecyl sulfate (final concentration, 3.0%) solution, and approximately 0.3 g of a mixture of glass beads that consisted of equal weights of (i) 0.1-mm- and (ii) 1-mm-diameter beads in a 2.5-ml polypropylene tube. The mixture was then vigorously shaken at 4,500 rpm for 5 min on a Micro Smash MS-100 apparatus (Tomy Seiko Co., Ltd., Tokyo, Japan), and the supernatant was collected by centrifugation at 20,000 × g for 5 min at room temperature. This DNA extraction was carried out three times. The three supernatants were combined and treated with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol). The DNA in the aqueous phase was concentrated and replaced by about 30 μl of TE buffer using Montage PCR centrifugal filter devices (Millipore, Bedford, MA). Using the extracted DNA as a template, the partial 16S rRNA gene fragments (approximately 580 bp) were amplified by a PCR method with a pair of universal primers (341F [5′-CCTACGGGAGGCAGCAG-3′] and 907R [5′-CCGTCAATTCMTTTRAGTTT-3′]) (5). Cycling conditions were 96°C for 5 min, followed by 30 cycles of 96°C for 30 s, 53°C for 30 s, and 72°C for 1 min, with a final elongation step at 72°C for 7 min, with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Foster City, CA). The amplified products were cloned into Escherichia coli using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Nucleotide sequences of the randomly chosen 58 clones were determined using the BigDye Terminator v3.1 cycle sequencing kit with the ABI3130xl sequencer (Applied Biosystems, CA). Homology searches were performed with the Basic Local Alignment Search Tool (BLAST) using an in-house software system and database, which collected type strains from the Ribosomal Database Project II (http://rdp.cme.msu.edu/).
The bacterial composition of the BALF can be found in Table 1. Twenty-eight of 58 clones (48.3%) were the most similar to the Leptotrichia wadei sequence reported under GenBank accession number AY029802 (97 to 96% homology). Ten clones (17.2%) were Veillonella spp. (five clones were the most similar to V. parvula sequence reported under GenBank accession number X84005 [99 to 97% homology], three were the most similar to the V. atypica sequence reported under accession number X84007 [99% homology], and two were the most similar to the V. dispar sequence reported under accession number X84006 [99% homology]). Seven clones (12.1%) were Enterococcus spp. (six clones were the most similar to the E. casseliflavus sequence reported under GenBank accession number Y18161 [99 to 92% homology], and one clone was the most similar to the E. faecalis sequence reported under accession number AB012212 [99% homology]). Five clones (8.6%) were Prevotella nanceiensis (GenBank accession number EF405529) (100 to 98% homology). These results indicated that Leptotrichia sp. was dominant in the BALF. Typical morphological descriptions of the genus Leptotrichia were compatible with those of the outnumbering bacteria observed in the Gram and Giemsa stainings. Veillonella spp. and Enterococcus sp. were also microscopically observed as gram-negative and -positive cocci, respectively. Overall, the bacterial composition analyzed by the molecular method was highly compatible with the microscopic findings. These findings suggested that Leptotrichia played relevant roles in this pneumonia together with mixed oral bacteria.
TABLE 1.
Bacterial composition of the clone library from the BALF specimen
| Organism (GenBank accession no.) | No. of clones | % Homologya
|
|
|---|---|---|---|
| Max | Min | ||
| Enterococcus faecalis (AB012212) | 1 | 99 | 99 |
| Enterococcus casseliflavus (Y18161) | 6 | 99 | 92 |
| Veillonella parvula (X84005) | 5 | 99 | 97 |
| Veillonella atypica (X84007) | 3 | 99 | 99 |
| Veillonella dispar (X84006) | 2 | 99 | 99 |
| Leptotrichia wadei (AY029802) | 28 | 97 | 96 |
| Prevotella nanceiensis (EF405529) | 5 | 100 | 98 |
| Otherb | 8 | ||
| Total | 58 | ||
Max and Min indicate the highest and the lowest similarities between the nucleotide sequence of the cloned fragment and the 16S rRNA gene sequence of each type strain, respectively.
Other organisms were Streptococcus spp. (three clones), Delftia sp. (one clone), Lactobacillus sp. (one clone), Syntrophococcus sp. (one clone), Clostridium sp. (one clone), and Actinomyces sp. (one clone).
Before we obtained the results of the molecular analysis, the patient had been treated by three antibacterial drugs having wide spectrums (such as imipenem-cilastatin [0.5 g twice per day], minocycline [100 mg twice per day], and sulfamethoxazole-trimethoprim [3 g four times per day]) from the day of admission to the seventh day in our hospital. The patient then slowly showed signs of improvement; he no longer required O2 supply, leukocytosis improved, and CRP levels decreased (CRP, 9.9 mg/dl). On the basis of the molecular information on the seventh day, the treatment was changed as follows: sulfamethoxazole-trimethoprim and minocycline were discontinued on the 7th day, imipenem-cilastatin was continued until the 10th day, clindamycin was administrated as an antianaerobic agent from the 7th day to the 19th day, and the patient was then discharged on the 30th day, with a complete recovery.
To determine whether Leptotrichia spp. were in his mouth or not, bacterial flora of gargled water was also analyzed. The sample was obtained just after the antimicrobiotic treatments were discontinued. The analysis revealed a total of 75 clones. Forty-five clones (60.0%), 9 clones (12.0%), 8 clones (10.7%), and 7 clones (8.6%) were the most similar to Staphylococcus sp., Enterococcus spp., Lactobacillus spp., and Acidaminococcus sp., respectively. We could not find any DNA sequences similar to those of Leptotrichia species by analysis of the gargled water.
This is the first report of pneumonia potentially caused by Leptotrichia sp. as being a major responsible agent in a healthy subject. Leptotrichia spp. are anaerobic rods and a constituent of the normal flora of the oral cavity (4), and they have rarely been reported to cause serious infectious diseases (8, 10, 11). In the past, some serious infectious diseases due to Leptotrichia spp. have been reported, such as bacteremia, endocarditis, hepatic abscess, and genital tract infection in immunocompromised patients or those with poor oral hygiene, during the perinatal period, or with cardiovascular abnormalities (1, 7-11).
We think that there are two explanations for the rarity of Leptotrichia infection. First, it is difficult to cultivate anaerobes. The anaerobic cultivation method is complicated and time-consuming (approximately 1 week or more). Second, we sometimes pass over Leptotrichia species as a real causative agent because it is a member of oral bacterial flora. In cases of mixed infection by indigenous or less virulent bacteria, it is critical to distinguish whether they are only contaminants or actual causative agents. These two problems may sometimes delay accurate diagnosis and adequate treatment.
In this case, we found the indigenous oral bacteria to be a possible responsible pathogen by microscopic findings and molecular analysis of the BALF sample. There is a small possibility that the specimen was contaminated by the bacteria of the oropharyngeal flora. However, it was previously reported that oral bacterial flora rarely affects or contaminates BALF specimens obtained by fiberoptic bronchoscopy (6). Because BALF specimens are obtained directly from the infected area of pneumonia, it is highly possible that the bacteria derived from BALF are the responsible agents. In the present case, the bacterial composition of gargled water that was obtained (just after discontinuation of antibiotic administration following successful treatments) did not contain Leptotrichia sp. These results strongly suggest that the bacterium was not just a contaminant from normal oral flora but played significant roles in this infection.
Some molecular methods using broad-range PCRs of the 16S rRNA gene have been applied to clinical practice to detect causative agents of infectious diseases (2, 3). Theoretically, they are designed to detect almost any causative bacteria. This may help clinicians to detect uncultivable bacteria due to fastidious cultural conditions or prior empirical antibiotic treatments. In addition, they may be able to identify pathogens that have never been recognized as a real etiological agent. In this case, Leptotrichia sp. as a potentially major responsible bacterium for severe pneumonia was detected by cultivation-independent methods. Based on these findings, the patient in the present case was successfully treated by monotherapy of clindamycin.
In conclusion, clinicians should be aware of (i) the possibility that Leptotrichia spp. may be related to serious pneumonia in both immunocompromised and immunocompetent patients and (ii) the potential of 16S rRNA gene sequencing analysis in a clinical setting.
Footnotes
Published ahead of print on 3 December 2008.
REFERENCES
- 1.Duperval, R., S. Beland, and J. A. Marcoux. 1984. Infective endocarditis due to Leptotrichia buccalis: a case report. Can. Med. Assoc. J. 130422-424. [PMC free article] [PubMed] [Google Scholar]
- 2.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 3531899-1911. [DOI] [PubMed] [Google Scholar]
- 3.Harris, K. A., and J. C. Hartley. 2003. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. J. Med. Microbiol. 52685-691. [DOI] [PubMed] [Google Scholar]
- 4.Hofstad, T. 1998. Fusobacterium and Leptotrichia, p. 1355-1364. In L. Collier, A. Balows, and M. Sussman (ed.), Topley and Wilson's microbiology and microbial infections, 9th ed., vol. 2. Systematic bacteriology. Arnold, London, United Kingdom. [Google Scholar]
- 5.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebraundt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., New York, NY.
- 6.Liebler, J. M., and C. J. Markin. 2000. Fiberoptic bronchoscopy for diagnosis and treatment. Crit. Care Clin. 1683-100. [DOI] [PubMed] [Google Scholar]
- 7.Messiaen, T., C. Lefebvre, and A. Geubel. 1996. Hepatic abscess likely related to Leptotrichia buccalis in an immunocompetent patient. Liver 16342-343. [DOI] [PubMed] [Google Scholar]
- 8.Morgenstein, A. A., D. M. Citron, B. Orisek, and S. M. Finegold. 1980. Serious infection with Leptotrichia buccalis. Report of a case and review of the literature. Am. J. Med. 69782-785. [DOI] [PubMed] [Google Scholar]
- 9.Shukla, S. K., P. R. Meier, P. D. Mitchell, D. N. Frank, and K. D. Reed. 2002. Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J. Clin. Microbiol. 403346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulstrup, A. K., and S. H. Hartzen. 2006. Leptotrichia buccalis: a rare cause of bacteraemia in non-neutropenic patients. Scand. J. Infect. Dis. 38712-716. [DOI] [PubMed] [Google Scholar]
- 11.Vemelen, K., I. Mertens, J. Thomas, J. Vandeven, J. Verhaegen, and L. Verbist. 1996. Bacteraemia with Leptotrichia buccalis: report of a case and review of the literature. Acta Clin. Belg. 51265-270. [DOI] [PubMed] [Google Scholar]