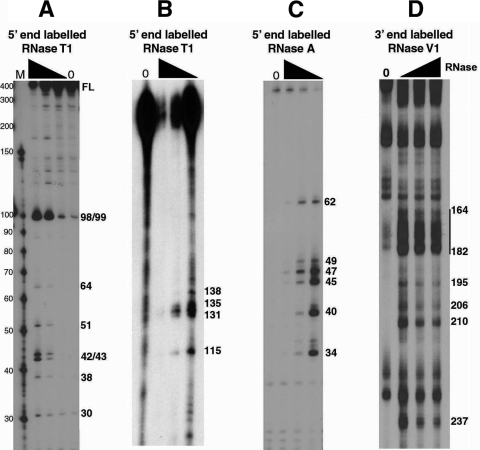
FIG. 1.
Partial RNase digestion shows single- and double-stranded regions of RSE RNA. (A and B) The 5′-end-labeled RSE RNA (294 nt) was digested with increasing amounts of RNase T1. The resulting cleavage products were run on a 15% (A) or 8% (B) polyacrylamide-8 M urea gel. Note that in panel A the 294-nt RSE full-length (FL) RNA comigrates with a marker of approximately 400 nt. (C) The 5′-end-labeled RSE RNA was digested with increasing amounts of RNase A and resolved on an 8 M urea-15% polyacrylamide gel. (D) The 3′-end-labeled RSE RNA was digested with RNase V1 and resolved on an 8 M urea-8% polyacrylamide gel. Numbers on the right correspond to the RSE nucleotide that is cleaved by RNase. Nucleotide position 4 of the RSE RNA corresponds to nt 2597 of the complete RSV genomic RNA sequence. The black triangle indicates increasing RNase concentrations. Lanes 0, RNA alone (no RNase added).