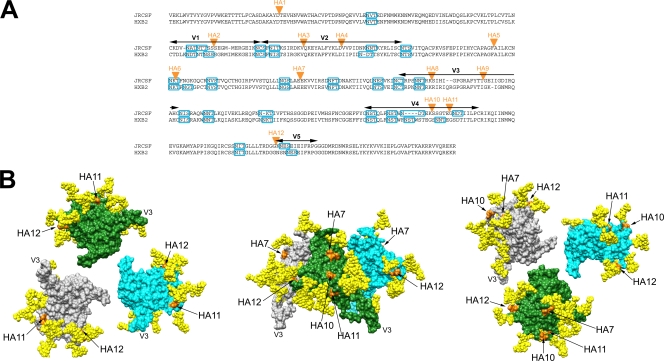
FIG. 2.
Locations of HA epitope tag insertions in JRCSF and HXB2. (A) Protein sequence alignment of JRCSF and HXB2 gp120s, denoting the locations of HA epitope tag insertions (orange triangles). The sequence alignments were made with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Sections of variable regions in gp120 are marked by arrows (black), and potential N-glycosylation sequons (PNGS) are denoted by rectangular boxes (blue). JRCSF contains 23 PNGS and HXB2 24 PNGS. The locations of the HA tag insertions are as follows: HA1, C1 region; HA2, V1 region; HA3 and HA4, V2 region; HA5, HA6, and HA7, C2 region; HA8 and HA9, V3 region; HA10 and HA11, V4 region; HA12, V5 region. (B) Surface representation of an unliganded gp120 spike from three different perspectives, illustrating the locations of the HA tag insertions. Left panel, from the perspective of the target cell, with V3 projecting toward the viewer. Middle panel, side view of the unliganded trimer, with the V3 loops pointing downward to the target cell and the virus surface at the top of the image. Right panel, from the perspective of gp41/virion surface, with V3 projecting away from the viewer. Each gp120 protomer (derived from gp120coreJRFL+V3; PDB ID 2B4C) is depicted in a different color (gray, green, or cyan). The oligomer configuration is based on cryo-electron microscopy images of an unliganded gp120 trimer fitted with X-ray coordinates of the gp120core+V3 structure (37). The GlcNAc2Man3 pentose cores of glycans attached to the Asn glycosylation site in PNGS are shown as space-filling models (yellow). The GlcNAc2Man3 cores were modeled using GlyProt (4). We modeled only the pentose core, given that it is the same in high-mannose, complex, and hybrid glycans that occur naturally on gp120. The two gp120 residues between which the HA epitope tag was inserted are colored orange; the locations of HA2 and HA3 cannot be depicted because the V1/V2 loop is truncated in this and all gp120 crystal structures. The molecular graphic images were generated with the UCSF Chimera package (54) and then labeled and compiled in Adobe Photoshop CS (version 8.0).