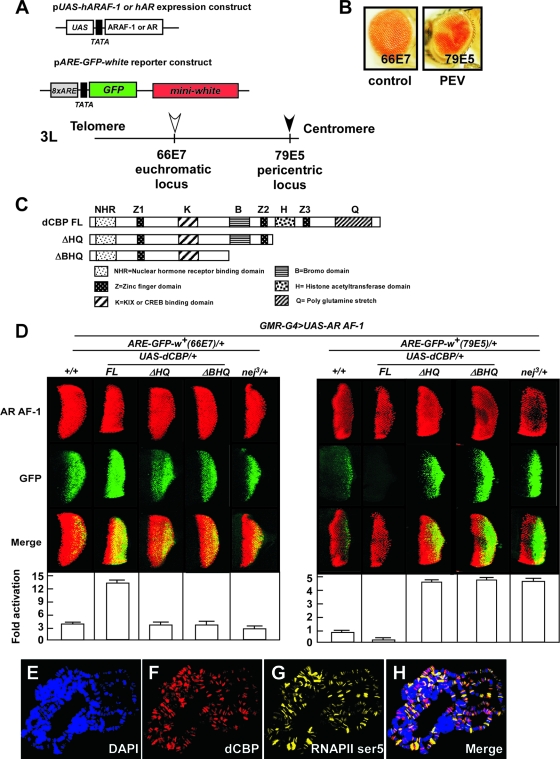
FIG. 1.
dCBP corepresses AR AF-1-induced transactivation at the pericentric region. (A) Schematic representation of the expression and reporter constructs and cytogenetic location of the integrated reporter gene in the chromosome in newly generated PEV fly lines. The expression constructs include the human AR or ARAF-1 driven by a UAS promoter. The reporter construct harbors the GFP reporter gene controlled by eight ARE copies and the white reporter gene driven by its endogenous promoter. (B) The eye of a novel PEV fly carrying the reporter transgene (ARE-GFP-white) inserted at a pericentric region by P-element mobilization and indicated as a filled arrow in panel A (79E5) shows a mosaic phenotype (right panel). In contrast, the control fly carrying a euchromatic transgene, indicated as an open arrow in panel A (66E7), has a uniformly red eye (left panel). The insertion sites were confirmed by inverse PCR. (C) Diagram of constructs for the full-length dCBP (dCBP FL) and dCBP truncation mutants. dCBP ΔHQ was used to produce the truncated HAT domain and mutations lacking a glutamine-rich stretch; dCBP ΔBHQ was used to produce the truncated bromodomain, HAT domain, and mutations lacking a glutamine-rich stretch. Each dCBP mutant was expressed within the developing eye by the use of a UAS-GAL4 misexpression system. (D) dCBP corepresses AR AF-1-induced transactivation at the pericentric area. Fly lines carrying a gain-of-function mutation of dCBP (GMR-G4/+; UAS-dCBP FL/+), truncation mutations (GMR-G4/+; UAS-dCBP ΔHQ/+ and GMR-G4/UAS-dCBP ΔBHQ), or a loss-of-function mutation (nej3/+) were crossed to two fly models expressing AR AF-1 proteins and harboring a ARE-GFP-white reporter gene in the euchromatic or pericentric region. Expression of the AR AF-1 controlled by a GMR-GAL4 (GMR-G4) driver in the UAS-GAL4 system in the third instar larvae eye imaginala discs was assessed by immunostaining using an anti-AR antibody (upper panels). The effects of dCBP and dCBP mutations on AR AF-1-mediated transactivation were assessed by examination of GFP expression (middle panels). Merged images are shown in the lower panels. Quantification of GFP expression revealed by color intensity gradations with Adobe Photoshop (histogram) is shown at the bottom. Error bars indicate standard deviations. (E to H) Polytene chromosomes from the third instar larvae of wt flies were dissected and stained with rat polyclonal anti-dCBP antibodies (red [F]) and mouse immunoglobulin M anti-RNAPII ser5 antibodies (yellow [G]) and with DAPI to visualize DNA (blue [E]). H, merged images. dCBP is ubiquitously present on the polytene chromosome, though it is weakly stained at the centromere. RNAPII ser5 and dCBP only partially merge, revealing the presence of dCBP in nontranscribed regions.