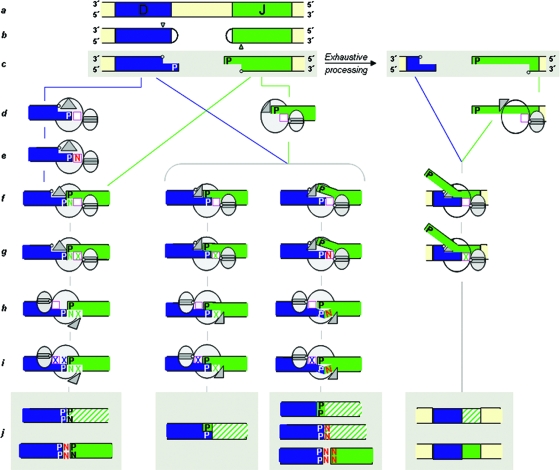
FIG. 6.
A model for nucleotide insertions catalyzed by Polμ during embryonic DJH rearrangements. The Polμ structure (in gray) is depicted as two ellipses, the large one representing the polymerization core and the small one indicating the 8-kDa domain, which has a single-stranded DNA binding cleft for the 5′ P binding site. The triangle represents mobile loop 1 of Polμ (39, 56). The deoxynucleotide triphosphate (dNTP) binding site is represented by a square (magenta). The hatched areas are “out-of-frame” J segments. (a) Germ line genomic region with D and J segments of the IgH chain. (b) RAG-induced hairpinned coding ends at both D and J segments. A nicking site is indicated by a small triangle at each end. (c) After nicking by Artemis-DNA-PK, a 3′ overhang (2 nt in this case), which contains a P nucleotide, is generated at both ends. An internal 5′ phosphate is indicated by a small circle. (d) Binding of Polμ to DNA ends can occur in two different ways: an enzyme-DNA complex can be formed in which the 3′ protruding end is oriented as a primer strand whose primer terminus (P nucleotide) is close to the dNTP binding site (left); alternatively, the 5′ P of one DNA end is bound by the Polμ 8-kDa domain next to the dNTP binding site (center and right). (e to j, left) Terminal-transferase N addition occurs before end joining (e). The inserted N nucleotide can serve as a connector for end joining if it is complementary to the other DNA end and the synapsis is stabilized by loop 1 (f). The N nucleotide, which is now inapparent as a template-independent insertion, is further extended by insertion of a templated dXTP (g). Once this strand is ligated, Polμ is adjusted to the remaining gap, coupled to a conformational change of loop 1 (h). After gap filling and ligation (i), the end joining is completed and produces an “out-of-frame” J segment (hatched area) due to the two extra P nucleotides added at the junction. An “in-frame” product (shown below) could be obtained by the addition of two N nucleotides before the end-joining step. In this case, one of the Ns would be evident (j). (f to j, middle left) Polμ's loop 1-mediated synapsis without requiring N nucleotides. If the two P nucleotides are complementary, their extension (X) will be template directed at both gaps, and the resulting product will not include N nucleotides (an “out-of-frame” J segment; one P nucleotide added). (f to j, middle right) If the two P nucleotides are not complementary and the templating base (located in front of the dNTP binding site) is not properly adjusted, Polμ can add N nucleotides. One or more N nucleotides could be immediately ligated and detected in the final recombination product. (c to j, right) When the ends are heavily processed and have large single-stranded overhangs, Polμ can bind the internal 5′ P and attract a second 3′ overhang to its vicinity, based on its capacity to accept microhomologies. After gap filling and further processing of the flapped strand, a largely deleted product is obtained that could contain either “out-of-frame” or “in-frame” J segments.