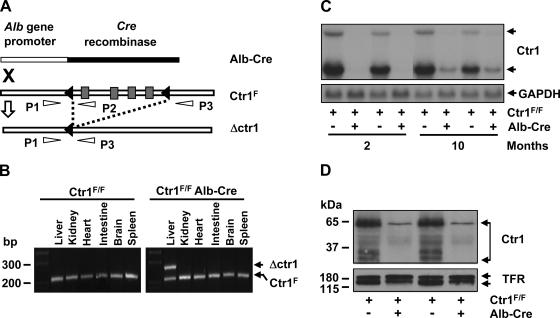
Fig. 1.
Deletion of Ctr1 in the liver. A: schematic depiction of chromosomal deletion of Ctr1 by recombination. Black triangles indicate loxp recombination sites that flank the four coding exons of Ctr1 (gray rectangles). A recombination event catalyzed by a Cre recombinase is indicated as dashed lines. P1, P2, and P3 indicate primers used for PCR genotyping of the loci. “X” indicates the intercross of mice possessing Ctr1F/F or a Cre recombinase gene fused with the albumin gene promoter. B: PCR analysis of recombination. Genomic DNA samples extracted from the organs of mice at 2 mo of age were subjected to PCR using P1, P2, and P3 primers. The fragments of 241 bp (PCR product of P1/P2 primer) and 281 bp (PCR product of P1/P3 primer) reflect Ctr1F and Ctr1 deletion (Δctr1), respectively. C: Northern blot analysis of Ctr1 transcripts. Total hepatic RNA isolated from 2 independent mice (1 male and 1 female) of each genotype was subjected to Northern blotting. A 32P-labeled DNA probe specific to mouse Ctr1 was used to detect mouse Ctr1 mRNA. A GAPDH-specific probe was used as a loading control as shown in the bottom panel. D: determination of Ctr1 protein expression by immunoblotting. Membrane proteins extracted from the liver of 2-mo-old mice were subjected to Western blot analysis of Ctr1 using anti-Ctr1 antibodies. Transferrin receptor (TFR) was probed as a loading control.