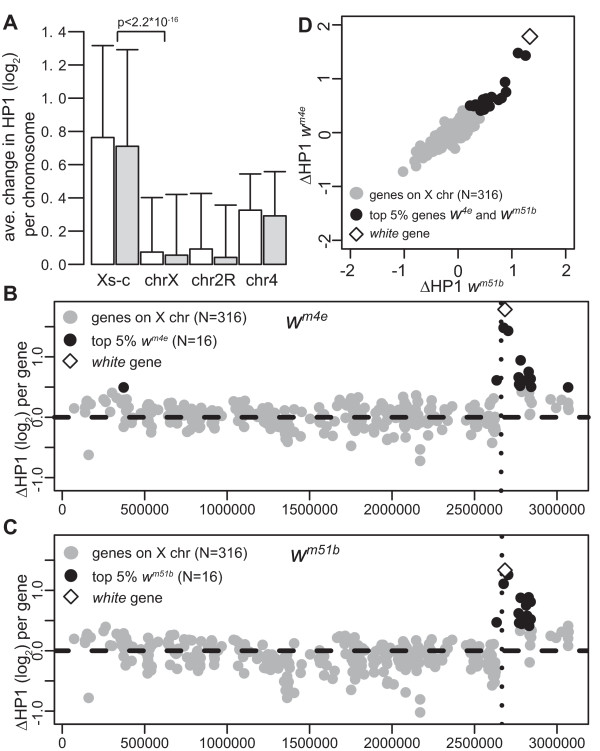
Figure 3.
Effects of wm inversions are found locally on X. A) Barplot showing the difference in HP1 binding for each probe on the array averaged per chr. Open bars show difference in binding between wm4e and wild-type, grey bars between wm51b and wild-type. Error bars indicate standard deviation of differences. Xs-c = XSyx4-CG3603; chrX = first 3.2 Mb of chr X excluding XSyx4-CG3603; chr2R = regions of chr 2R that are covered on the microarray; chr4 = chr 4. P value from Wilcoxon rank sum test. B-C) Chromosomal maps of first 3.4 Mb of X chr, with ΔHP1 (average change in log2 HP1 binding ratio) per gene. ΔHP1 between wm4e and Oregon-R-S (B), ΔHP1 between wm51b and Oregons-R-S (C). Black dots show top 5% of genes (n = 16) for which ΔHP1 is largest. Inversion breakpoints are indicated with black dotted lines. D) Bivariate scatterplot of data presented in B and C.