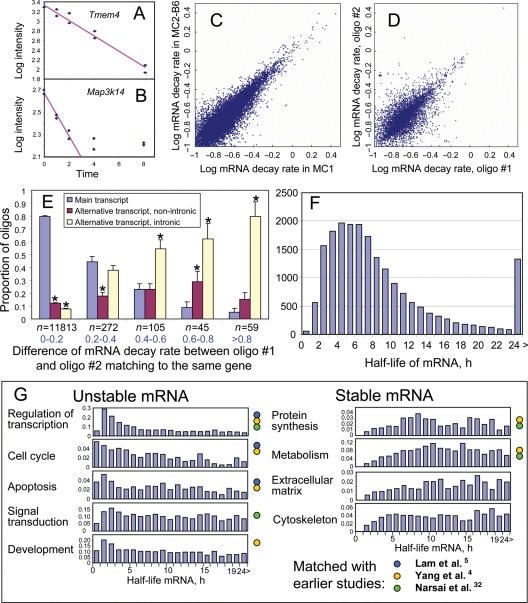
Figure 1.
Stability of mRNA in mouse ES cells. (A and B) Regression of gene expression level (microarray signal intensity in log scale) versus time after suppressing transcription by actinomycin D (y = a + bx) is used to estimate the rate of mRNA decay, d = b*ln(10). All 10 data points are used for an example (A) and only six data points are used for and example (B). (C) Comparison between mRNA decay rates estimated for MC1 ES cells and those estimated for MC2-B6 ES cells. Rates are log-transformed, log10(d + 0.1). (D) Comparison between mRNA decay rates estimated using different probes (#1 and #2) on the microarray, which match to the main transcript of the same gene. Only the data for MC1 cells are shown. (E) Comparison of mRNA decay rates identified with the best probe (#1) from the main transcript of each gene with decay rates estimated using other probe (#2) that matched the same gene; pairs of probes were first classified into groups according to the difference in estimated mRNA decay rate (group size is shown below bars, n), and then the proportion of probes #2 that matched different types of transcripts and/or introns was estimated and plotted for each group. (F) Frequency distribution of mRNA half-life among all genes. (G) Frequency distribution of mRNA half-life in different functional categories of genes; color circles indicate that the functional group was previously identified as being over-represented among stable or unstable mRNA species.