Figure 4.
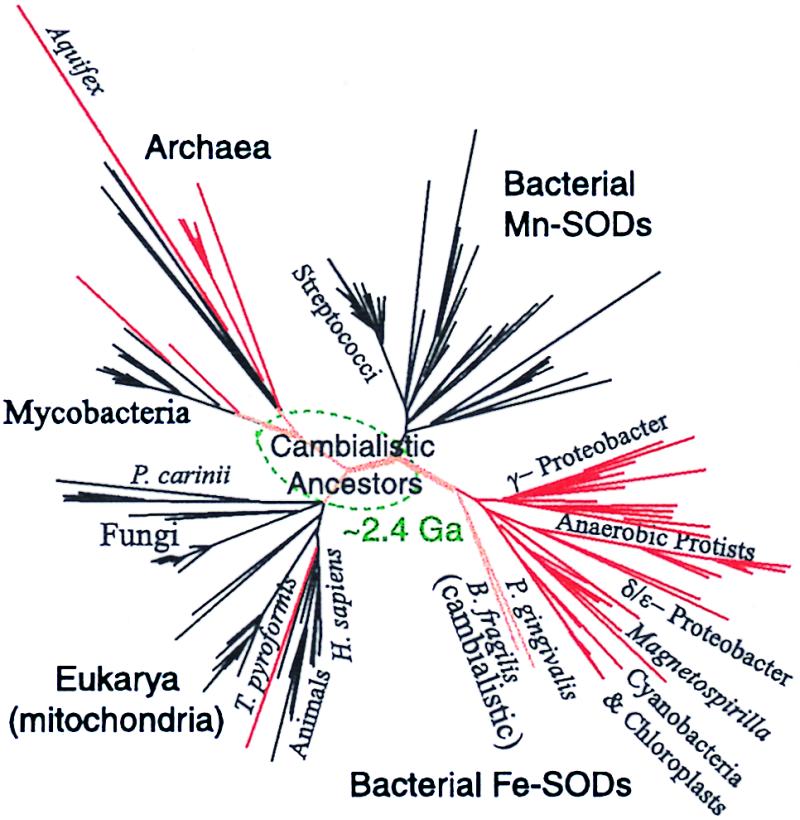
Unrooted phylogenetic tree constructed from the alignment of 214 Fe/Mn super oxide dismutase (SOD) amino acid sequences. The branches are color coded as Fe-SODs (red), cambialistic (Fe or Mn) SODs (orange), and Mn-SODs (black). The portion of the tree estimated to be older than the Paleoproterozoic snowball Earth event at 2.4 Ga is located inside the dotted oval, and hypothesized to be cambialistic. The National Center for Biotechnology Information protein and nucleotide database Entrez search tool was used to obtain 206 sequences; the sequences of two Magnetospirillum strains, MS-1 (ATCC 31632) and AMB-1 (ATCC 700264) and a Magnetovibrio strain MV-1 (gift of D. Dean, Virginia Tech.) were obtained from genomic DNA by PCR using the degenerate primers 5"-AIGTAGTAIGCITGITCCCA-3" and 5"-AIACIATGGAAATCCACCA-3". Additional sequence for MV-1 was obtained by PCR using 5"-AAGCACCACGIGICCTACGT-3" together with the second primer given above and for MS-1 by using the PCR product to probe a small insert EcoRI library of MS-1 DNA. An Aquifex sequence was obtained from ref. 64. The sequences were trimmed to a region similar to that chosen by Smith and Doolitle (53) of about 136 aa (in human) from just N-ward of the first His metal ligand to just C-ward of the Asp ligand. Multiple sequence alignment was performed by clustal w (65) using the blosum62 matrix (66) and was manually inspected by location of the extremely conserved metal ligand residues (His-His-Asp-His). The tree shown is heuristically generated by clustal w, and branch lengths are corrected for multiple substitutions. Although the precise position of many species is uncertain, particularly those with long branch lengths, the basic branching topology agrees with parsimony trees generated by using the phylip package (67) and is strongly supported by bootstrap analysis.
