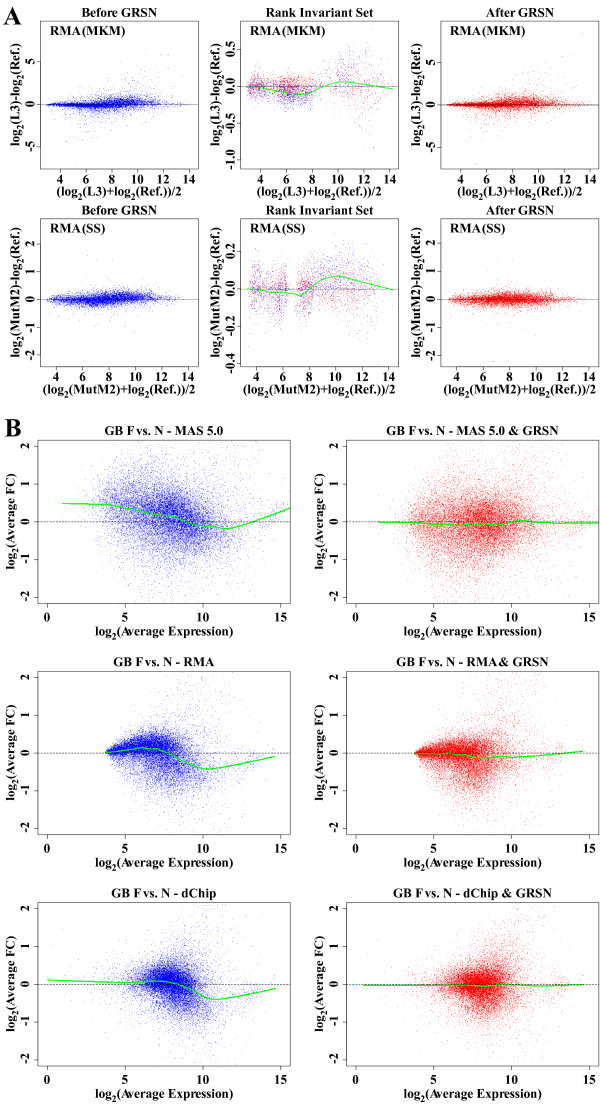
Figure 5.
GRSN corrects non-linear artifacts in representative microarray datasets. A. GRSN applied to two different microarray datasets. First row – late stage sample L3 from the MKM dataset. Second row – mutant Male sample MutM2 from the SS dataset. Columns 1–3 demonstrate the effect of GRSN on the selected samples as described in figure 2 above. The RMA probe set summary method was used in each. B. GRSN can reduce systematic non-linear artifacts which can affect fold change analysis regardless of pre-processing method. M vs. A plots showing fold change as a function of mean value and plotted on log base 2 scale. Both fold change and mean are calculated using multiple replicates, 14 FA samples and 11 Normal samples from the GB dataset (not just comparing two samples). A lowess smoothed curve is displayed to show the trend of the scatter plots. Three different summary methods are shown: Top row – MAS 5.0, Middle row – RMA, and Bottom row – dChip®. The results in the left column are without GRSN applied and the effect of applying GRSN to each of the respective methods is shown in the right column.