Abstract
The blood-brain barrier (BBB) is a vascular endothelial interface that separates the brain interior from the bloodstream. Membrane proteins resident at the BBB play important functional and regulatory roles. The current study describes the development and successful implementation of a multiplex expression cloning (MEC) method to allow facile identification of BBB membrane proteins. The overriding goal of the MEC approach was to mine a BBB cDNA library and selectively isolate membrane protein-encoding cDNAs. This selection process was achieved via fluorescence activated cell sorting (FACS) of cDNA-expressing mammalian host cells for those cells that were immunolabeled with a BBB membrane protein-specific polyclonal antiserum (BMSPA). After optimization of the host cell expression system, four selection rounds allowed the isolation of a panel of 15 unique cDNAs that encoded BBB membrane proteins. The identified proteins display significant diversity in structure, function and in vivo expression levels. The MEC approach thus proved effective for conducting moderate throughput membrane proteome analyses of the BBB while limiting any biases caused by membrane protein insolubility or low in vivo expression levels that can complicate other proteomic approaches.
Keywords: Blood-brain barrier, brain endothelial cells, brain microvasculature, expression cloning, membrane proteomics
1. INTRODUCTION
The brain microvasculature, also known as the blood-brain barrier (BBB), is composed of specialized endothelial cells that serve as an interface between the blood and the brain. As such, the BBB plays the role of regulating transport into and out of the brain while also defending the brain against circulating toxins. Though important for the healthy functioning of the brain, the BBB has also been implicated in the progression of several neurological disorders. In light of these facts, it is important to map the BBB membrane proteome as it helps dictate many of the specialized transport and signaling characteristics. Such an effort would help to better understand the role of the BBB in healthy individuals and may also aid in the identification of drug targets for the treatment of neurological disorders [1]. Several studies have already focused on comprehensive elucidation of the rat and human BBB transcriptome [2, 3, 4, 5]. While enlightening, these genomic studies do not directly reveal information about a protein's cellular localization or take into account the effect of translational and post-translational regulation issues on protein expression, thus highlighting a need for direct proteomic studies.
Membrane proteins tend to be insoluble due to their hydrophobic nature and can be heterogeneous in size as a result of post-translational modifications, such as glycosylation. Since current proteomic techniques, such as mass spectrometry (MS) and 2D gel analyses rely on solubilization and/or size resolution to sample and separate proteins, membrane proteomics poses a formidable challenge. Although rapid advances made in the field of MS [6] coupled with innovative sample preparation techniques have yielded an impressive coverage of membrane proteins [7, 8], instrumentation-based limitations such as sensitivity to contaminants, preference towards certain ion types [9], ionization suppression during electrospray ionization [10] and limitations on the number of peptides that can be co-eluted [9] can bias the results obtained from such studies. Thus a significant fraction of the membrane proteins that are highly insoluble and/or expressed in low abundance in vivo, although biologically relevant, may not be identified by such analyses [11].
As a complement to MS-based proteomic methods, we describe here a multiplex expression cloning (MEC) approach to help identify proteins that could be underrepresented in MS analyses. Previously, a BBB membrane protein-specific polyclonal antiserum (BMSPA) was used to help individually isolate three BBB membrane protein-encoding cDNAs from a BBB cDNA library [12, 13, 14]. In the current study this expression cloning method was combined with fluorescence activated cell sorting (FACS) to enable a moderate throughput analysis of the BBB membrane proteome. To help avoid potential biases resulting from membrane protein insolubility, the MEC method directly probes membrane proteins expressed in the near native context of a cell membrane. In addition, a constitutive CMV promoter and an SV40 origin of replication present on the BBB protein-encoding cDNAs ensure reasonably high protein expression levels in the host cell system regardless of protein abundance in vivo. Finally, MEC yields full-length cDNAs encoding for membrane proteins, which may be used not only for de novo identification of the protein but also for more focused individual protein characterization.
2. MATERIALS AND METHODS
2.1. Cell culture
COS-1 (green monkey kidney cell line) and HEK293 (human embryonic kidney cell line) cell lines were both obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured in a humidified incubator at 37°C with 5% carbon dioxide using DMEM media (Gibco, Invitrogen, Carlsbad, CA) buffered with 2.0 g/l sodium bicarbonate and 7.2 g/l HEPES to pH 7.2 and supplemented with 4.5 g/l dextrose and 10% fetal bovine serum (Gibco, Invitrogen).
2.2. BBB cDNA library and BMSPA
The BBB cDNA library was generated from bovine brain microvessels [12] and consists of about 5×106 BBB cDNA clones inserted into the PCDNA1.1 (Invitrogen) vector backbone. The PCDNA1.1 vector places the inserts under the control of a constitutive CMV promoter and possesses an SV40 origin of replication that allows episomal replication of the plasmid within a host cell that expresses the SV40 large T-antigen. The BMSPA was previously raised against isolated bovine brain capillary plasma membranes (kind gift of Dr. William Pardridge, Ref. [15]) and was incubated with an untransfected HEK293 (or COS-1) cell monolayer at 4°C for 1 hour to remove any background antigenicity (as seen in Figure 2D).
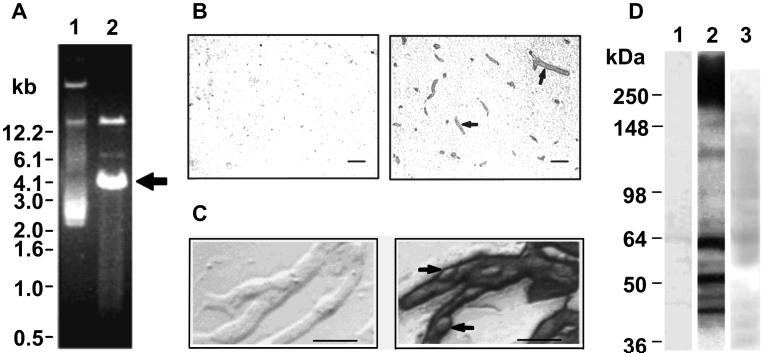
Figure 2.
A. Agarose gel analysis of the BBB cDNA library showing uncut plasmid pool (lane 1) and BamHI-XhoI restriction digested plasmid pool (lane 2); a bright band corresponding to the vector PCDNA1.1 can be seen at 4.0 kb (arrow). The cDNA inserts comprise a smear ranging from 0.5 to 12.2 kb. B. Bovine brain sections immunolabeled with the BMSPA (right) or pre-immune serum (left). Arrows indicate two examples of the many stained microvessels. Scale bar = 50 μm. C. Isolated microvessels labeled with BMSPA (right) or pre-immune serum (left). Arrows indicate lightly stained pericytes. Scale bar = 50 μm. D. Western blot of bovine capillary lysates separated by reducing SDS-PAGE and stained with the pre-immune serum (lane 1), the BMSPA (lane 2), or lysates of HEK293 cells transfected with no-insert PCDNA1.1 and probed with the BMSPA (lane 3).
2.3. Brain capillary isolation, tissue sectioning, immunostaining and western blotting
Bovine brain microvessels were isolated from total brain homogenates using a mechanical isolation procedure [12] and captured onto poly-L-lysine treated glass slides by 30 minutes of incubation at room temperature. Intact bovine brain tissue was embedded in a cryomold using Tissue-Tek O.C.T Compound (Sakura Finetek, Zoeterwoude, The Netherlands) and frozen by floating the cryomold in a bath of isopentane (Sigma Aldrich, St. Louis, MO) cooled by liquid nitrogen. Seven micron thick tissue sections were generated using a Microm HM505E (Microm GmbH, Walldorf, Germany) and placed onto glass slides. The capillaries or tissue sections were fixed in 100 % methanol (-20°C, 20 minutes), permeabilized in 250 mM EDTA (37°C, 20 minutes), immunolabeled (BMSPA dilution of 1:1000) and imaged on an Olympus I×70 microscope (Olympus America Inc., Center Valley, PA). Western blotting under reducing conditions was performed as described previously [12].
2.4. DNA preparation, amplification and analysis
Electrocompetent MC1061/P3 Escherichia coli cells were used to amplify and prepare all cDNA samples. The MC1061/P3 bacteria were transformed using a Genepulser instrument (Bio-Rad, Hercules, CA) set at 2500 V, 25 μF, 200 Ω and a 2mm-gap electroporation cuvette (BioRad). After one hour of incubation in SOC media (2% Bacto tryptone, 0.5 % Bacto yeast extract, 10 mM NaCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM dextrose) at 37°C, the transformation mix was plated on LB (1% Bacto tryptone, 0.5% Bacto yeast extract, 0.17 M NaCl) plates containing 10 μg/ml tetracycline and 50 μg/ml ampicillin and incubated overnight at 37°C. For MEC selections, recovered colonies were counted to determine the library diversity, and all bacterial clones were inoculated into liquid LB media as a single pool, amplified overnight at 37°C and the plasmid DNA was recovered using a miniprep kit (Qiagen, Valencia, CA). Where indicated, the relative amounts of BamHI (New England Biolabs, Beverly, MA) linearized plasmid DNA in each pool were quantified by image analysis of ethidium bromide stained agarose gels. For single cDNA clone analysis, plasmid DNA was recovered from individual bacterial colonies and sequenced using standard T7 forward and PCDM8 reverse primers. Identities of the cDNA clones were determined by using a BLAST search (NCBI) against the nucleotide collection (nr/nt) database for sequences derived from Bos Taurus.
2.5. Mammalian cell transfection
Transfection of COS-1 cells was performed using the optimized DEAE-dextran method exactly as described previously [12]. The HEK293 cells were transfected using a calcium-phosphate technique [16] optimized for MEC. Cells were plated 12-14 hours prior to transfection at a density of 8×104 cells / cm2, in tissue culture treated 24-well plates using 500 μl of media. The transfection mix was made by adding 25 μl of 50mM HEPES, 280 mM NaCl and 1.5 mM Na2HPO4 (pH 6.9) to a 25 μl solution consisting of 1.25 μg plasmid DNA and 3.125 μl 2M CaCl2 in ddH2O. The solution was allowed to stand at room temperature for 1 minute and then added dropwise to the well being transfected. For large scale transfections, T75 flasks were used and all values were appropriately scaled up for a plating volume of 15 ml instead of 500 μl. The cells were incubated in a humidified incubator at 37°C with 5% carbon dioxide for 60 hours to allow for protein expression and supplemented with fresh preheated (37°C) media 24 hours post-transfection. For imaging of immunolabeling of individual clones, the cells were instead transfected using Lipofectamine 2000 (Invitrogen) as per manufacturer's instructions. Lipofectamine 2000, while not suitable for expression cloning [12], achieves very high transfection efficiency (60-65% as compared to 25-30% for calcium phosphate for the Lutheran glycoprotein cDNA) and was hence used for imaging studies.
2.6. Immunolabeling for flow cytometric analyses & sorting
At 60 hours post-transfection, the culture medium was aspirated and the transfected monolayers were washed once (5 ml per T75) with 0.01 M PBS, pH 7.4 supplemented with 10% fetal bovine serum (PBSF). The cells were then blocked with 40% goat serum (Sigma Aldrich) in 0.01 M PBS, pH 7.4 (PBSG, 4°C, 30 minutes), washed once more in PBSF and incubated in HEK293 (or COS-1) depleted BMSPA (1:500 in PBSG, 4°C, 1 hour with gentle rocking). The cells were then washed twice in PBSF, incubated in HEK293 (or COS-1) depleted phycoerythrin (PE) conjugated anti-rabbit IgG (Sigma Aldrich, 1:200 in PBSG, 4°C, 30 minutes), and washed twice with PBSF. A non-enzymatic cell dissociation solution (Sigma Aldrich) supplemented with 10% fetal bovine serum was used to release the transfected cells from the culture surface. After centrifugation at 200g for 5 minutes, the supernatant was aspirated and the cells resuspended in PBSF for flow cytometric analysis.
Fluorescence activated cell sorting (FACS) was conducted on a FACSVantage SE cytometer (Becton Dickinson) at the UW Paul P. Carbone Comprehensive Cancer Center Flow cytometry core facility. Cells were gated to exclude permeabilized cells (as indicated by a nuclear DAPI stain) and doublets (based on forward scatter peak widths). The sorting gate was conservatively set to include cells having fluorescence just above that of mock-transfected cells (see Figure 5B). A modified Hirt's extraction method [12] was used to recover the plasmid cDNA pool from sorted HEK293 (or COS-1) cells.
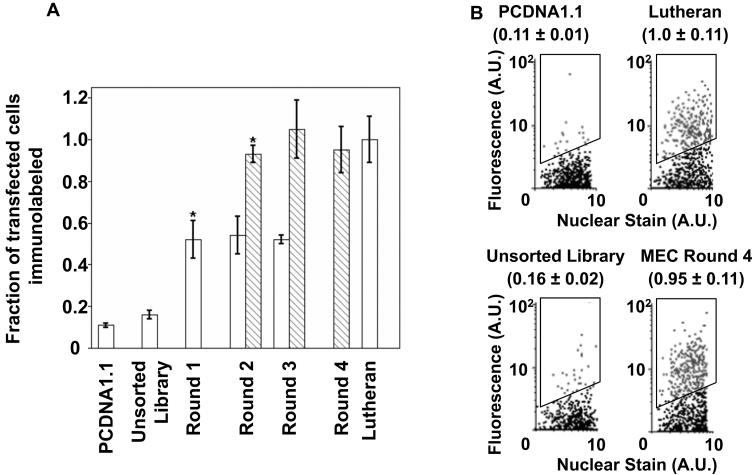
Figure 5.
Enrichment of cDNA library by MEC. A. Fraction of HEK293 cells that immunolabel with the BMSPA as defined by falling into the fluorescence sorting gate (illustrated in panel B). Values are normalized to that of the positive control, Lutheran cDNA-transfected cells (~25% of the total number of Lutheran-transfected cells fall into the sorting gate). Open bars indicate pools sorted without any plasmid dilution prior to transfection, whereas the striped bars indicate those that were sorted with a 100-fold dilution of cDNA pool prior to transfection. Error bars denote SEM for 3 measurements each; * = p<0.05 compared to previous MEC round as determined by a two-tailed Student's t-test. B. Flow cytometry dot plots indicating the fluorescent labeling of transfected HEK293 cells for some representative samples from Figure 5A. Black events (dots) indicate the non-transfected (or not recognized by BMSPA) cell population whereas the gray events (dots) indicate cells that fall within the cell sorting gate (outlined region). A conservative sorting gate was used to recover even those membrane protein-encoding cDNA that yield low fluorescence signals. Hence, some negative events (cDNA that do not encode membrane proteins, e.g. PCDNA1.1) also fall within the sort gate and are recovered in a given MEC round, but are subsequently lost due to the multiple round nature of the MEC process. Values in parentheses are consistent with Figure 5A, and indicate the fraction of transfected cells that are fall within the sort gate (gray dots), along with SEM. In all cases, the only cell populations evaluated and shown in the flow cytometric dot plots were those that exclude propidium iodide nuclear stain.
2.7. Immunocytochemistry
At 60 hours post-transfection, the lipofectamine 2000 transfected cells were washed once in PBSF, blocked in PBSG (150 μl per well, 4°C, 30 minutes), washed once in PBSF and incubated in the HEK293 depleted BMSPA (1:250 in PBSG, 4°C, 1 hour). After washing three times in PBSF, the cells were incubated with HEK293 depleted Alexa488 conjugated anti-rabbit IgG (Invitrogen, diluted 1:200 in PBSG, 4°C, 30 minutes), washed three times in PBSF. Cell imaging was performed on an Olympus IX70 fluorescence microscope using an excitation wavelength of 445 ± 20 nm and emission wavelength of 509 ± 24 nm.
3. RESULTS
3.1. Multiplex Expression Cloning (MEC) Strategy
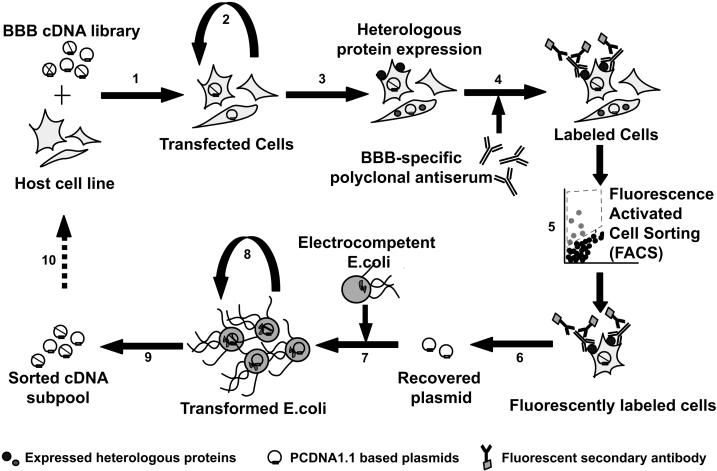
The schematic for the MEC approach is shown in Figure 1. As indicated, the BBB cDNA library [12] is first transfected into a mammalian host cell line such as COS-1 (Figure 1, step 1) for protein expression (Figure 1, step 3). The transfected plasmids contain an SV40 origin that allows episomal replication within the host cell line (Figure 1, step 2), facilitating increased protein expression and a better chance of recovering target cDNA later (Figure 1, step 6). It has been shown that the copy numbers for plasmids sized between 3-5 kb and containing the SV40 origin of replication can range from 2×105 to 4×105 per COS-1 cell [17]. Next, the mammalian host cells expressing the BBB membrane proteins are identified using the BMSPA [15] and a fluorescent secondary antibody (Figure 1, step 4). Importantly, other proteins encoded by the BBB cDNA library that are not expressed on the cell surface, such as cytosolic proteins, are generally not recognized by, nor accessible (given the live cell labeling approach) to the BMSPA. To recover the host cells expressing BBB membrane proteins, fluorescently immunolabeled cells are isolated from the diverse cell mixture via high throughput fluorescence activated cell sorting or FACS (Figure 1, step 5). The target plasmid cDNA pool is then purified from the isolated host cells via modified Hirt extraction (Figure 1, step 6) and subsequently amplified in E.coli (Figure 1, steps 7-9) to yield a new plasmid pool enriched for membrane protein-encoding cDNAs. This completes one “round” of selection. The newly generated cDNA pool can be subjected to subsequent rounds of MEC (Figure 1, step 10) until it is predominantly comprised of membrane protein-encoding clones that can be sequenced for gene identification.
Figure 1.
Schematic for MEC: 1. Transfection of host cell line, 2. Episomal replication of plasmids, 3. Protein expression, 4. Immunolabeling with BMSPA, 5. FACS, 6. Modified Hirt's extraction for plasmid recovery, 7. Plasmid electroporation into E. coli, 8. Bacterial amplification of enriched cDNA pool, 9. Preparation of enriched cDNA pool, 10. Repeat until highly enriched in BBB membrane protein-encoding cDNA.
The two key components necessary for the success of the MEC approach are the BBB cDNA library and the BMSPA. The BBB cDNA library consists of approximately 5×106 clones and was generated using mRNA recovered from freshly isolated bovine brain capillaries as described previously [12]. As shown in Figure 2A, the bovine BBB cDNA library is composed of a 4.0 kb vector backbone with cDNA insert sizes spanning from 0.6 to 12 kb. The cDNA inserts are placed under the control of a constitutive CMV promoter that drives increased expression of all proteins, irrespective of their in vivo expression levels. As demonstrated previously, the BMSPA, which was raised against intact brain microvessel plasma membrane sheets [15], selectively targets the blood vessels in the brain parenchyma (Figure 2B) and labels brain microvessel endothelium more intensely than pericytes (Figure 2C) indicating a reasonable selectivity for endothelial membrane proteins. The BMSPA was raised against isolated bovine brain capillary plasma membranes [15] and specifically recognizes a panel of putative BBB membrane proteins as indicated by Western blotting with brain microvessel lysates (Figure 2D), and the MEC approach was developed with the aim of determining the identity of these membrane proteins.
3.2. Validation of the MEC Approach Using Lutheran Glycoprotein
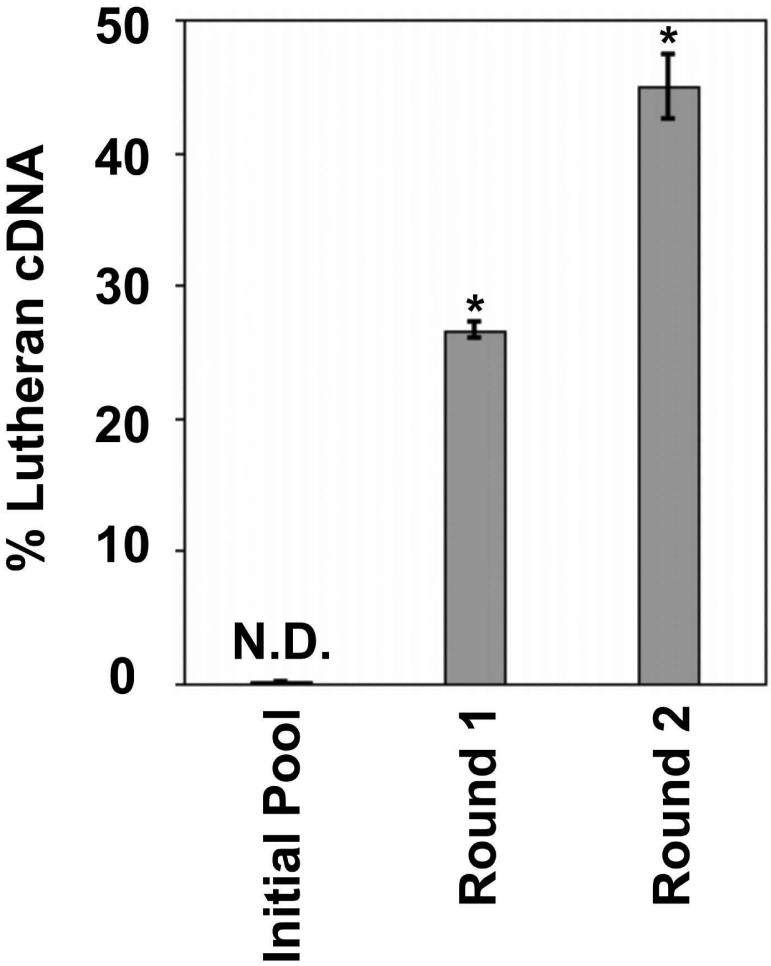
Previously, a manual expression cloning approach utilizing the cDNA library and the BMSPA described above led to the discovery of 3 membrane proteins expressed selectively at the bovine BBB [12, 13, 14]. Since this approach relied on manual identification and isolation of immunolabeled cells, it was far from a comprehensive screening of the membrane protein-encoding clones recognized by the BMSPA. The Lutheran glycoprotein, a glycosylated transmembrane protein that plays a role in basal cell adhesion, was one of the proteins identified in these studies and was therefore used as a positive control for the validation of the new FACS-based MEC approach. A plasmid pool consisting of Lutheran-encoding cDNA diluted 1:1000 in the no-insert PCDNA1.1 vector was generated. The recovery of Lutheran-encoding cDNA from this pool would be representative of the successful recovery of target cDNA from a background of the BBB cDNA library clones that are not membrane expressed or not recognized by the BMSPA. The pool was subjected to two rounds of MEC and the fraction of the Lutheran-encoding cDNA was determined by quantitative agarose gel electrophoresis. The percent of Lutheran-encoding cDNA increased from undetectable levels (i.e. 0.1%) in the starting pool to 45% of the enriched pool after just two rounds of sorting (Figure 3), indicating the successful recovery of a BBB membrane protein-encoding cDNA from a large background of irrelevant plasmids.
Figure 3.
Quantitative agarose gel analysis results for MEC enrichment of Lutheran glycoprotein-encoding cDNA from a 1:1000 dilution with no-insert PCDNA1.1. Error bars denote SEM for 4 independent agarose gel analyses each; N.D. = Not Detected; * = p<0.05 compared to previous round as determined by a two-tailed Student's t-test.
3.3. Implementation of the MEC Approach Using the BBB cDNA Library
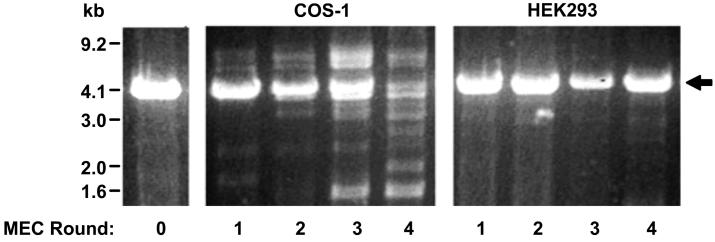
Efforts to apply the Lutheran-optimized MEC process to the actual BBB cDNA library led to the identification of several additional problems. As the number of MEC rounds increased (a necessary by-product for the enrichment of low frequency clones), there was a steady loss of full-length plasmid in the enriched cDNA pools as evidenced by the disappearance of the PCDNA1.1 backbone (Figure 4, COS-1 panel). Analyses of individual plasmids from these pools revealed the presence of “contaminating” DNA plasmids that possessed the antibiotic resistance gene, but lacked the full vector backbone of the BBB cDNA library (data not shown). Being relatively small in size, they amplified at faster rates than full-length BBB cDNA clones during each round of selection thereby counteracting the enrichment process. Closer inspection of each step in the MEC scheme revealed that the detrimental processing of plasmids in the COS-1 host cells themselves caused the appearance of these problematic plasmids (Figure 1, step 2). Indeed, COS-1 cells have been reported to have a relatively high frequency of mutation of heterologous plasmids; close to 2-3% of incoming plasmids are mutated with 72-80% of these being `deletion' mutations where entire components of the plasmids are deleted [18], a property that is particularly exacerbated by the multi-round nature of the MEC process. Alternatively, the HEK293 cell line, which is derived from human embryonic kidney cells [19], displays a mutation rate of 0.035-0.05%, with only 15-30% of these comprising deletion mutants [20]. In addition, the HEK293 cell line also supports episomal replication of SV40 origin-containing vectors at the same level as COS cells [21]. Thus, switching from COS-1 to HEK293 as the MEC host cell (Figure 1, step 1) resolved the plasmid contamination issue and no detectable loss in intact plasmid levels was observed even after four rounds of MEC using HEK293 cells (Figure 4, HEK293 panel).
Figure 4.
Agarose gel image displaying starting BBB cDNA library (left), cDNA pools subject to four rounds of MEC using COS-1 cells as the host (center) or HEK293 cells as the host (right). All lanes are BamHI-XhoI restriction digested to release the inserts. The PCDNA1.1 vector backbone is indicated with an arrow. Prominent high and low molecular weight bands appearing in COS-1 lanes 3 and 4 are contaminating plasmids.
Despite the change to HEK293 as the host cell line, three successive rounds of MEC failed to significantly enrich the BBB cDNA pool. The fraction of HEK293 cells transfected with the recovered cDNA pool that were fluorescently immunolabeled by the BMSPA initially increased to about half of total transfected cells after round 1, but failed to further increase in subsequent rounds (open bars, Figure 5A). The fraction of the fluorescently immunolabeled transfected HEK293 cells represents the probability of each cell sampling at least one membrane protein-encoding cDNA during the transfection process. However, each HEK293 cell likely uptakes multiple plasmids during transfection (Figure 1, step 1), implying that each individual cell isolated via FACS harbors several plasmids, only one of which is required for the cell to be fluorescently immunolabeled. Any further enrichment (Figure 1, step 5) of the target clones in subsequent MEC rounds is thus hampered. This hypothesis was corroborated by a plasmid titration study with the Lutheran-encoding cDNA, which revealed that each HEK293 cell uptakes an average of about 80 plasmids (data not shown). These data suggested that all transfected cells would label immunopositive even though only 1 in 80 plasmids would be a target membrane protein-encoding cDNA. To reduce the uptake of multiple independent BBB cDNA plasmids by a single cell during transfection (Figure 1, step 1), the BBB protein-encoding cDNA clones were diluted a 100-fold in a `dummy plasmid', puc18. The use of puc18 as a `dummy plasmid' maintains the overall transfection efficiency by keeping the total DNA concentration constant in the transfection mix, while providing the ability to eliminate puc18 from the recovered clones by appropriate antibiotic selection during the bacterial amplification step (Figure 1, step 8). As revealed by the striped bars in figure 5A, a 100-fold dilution prior to transfection led to further enrichment of the cDNA pool for membrane protein-encoding clones. Despite the apparent enrichment of the pool after a single additional round of selection using plasmid dilution (Figure 5A, striped bar for round 2), two further rounds were required in order to detect membrane protein-encoding cDNA clones at a reasonable frequency during individual clone analysis of the recovered pools. After the fourth round of MEC using the plasmid dilution approach, the frequency of membrane protein-encoding clones in the recovered cDNA pool increased from 1 in 80 to about 1 in 7. In principle, this enrichment could be further improved by subsequent rounds of MEC or a greater than 100-fold dilution of the plasmid cDNA.
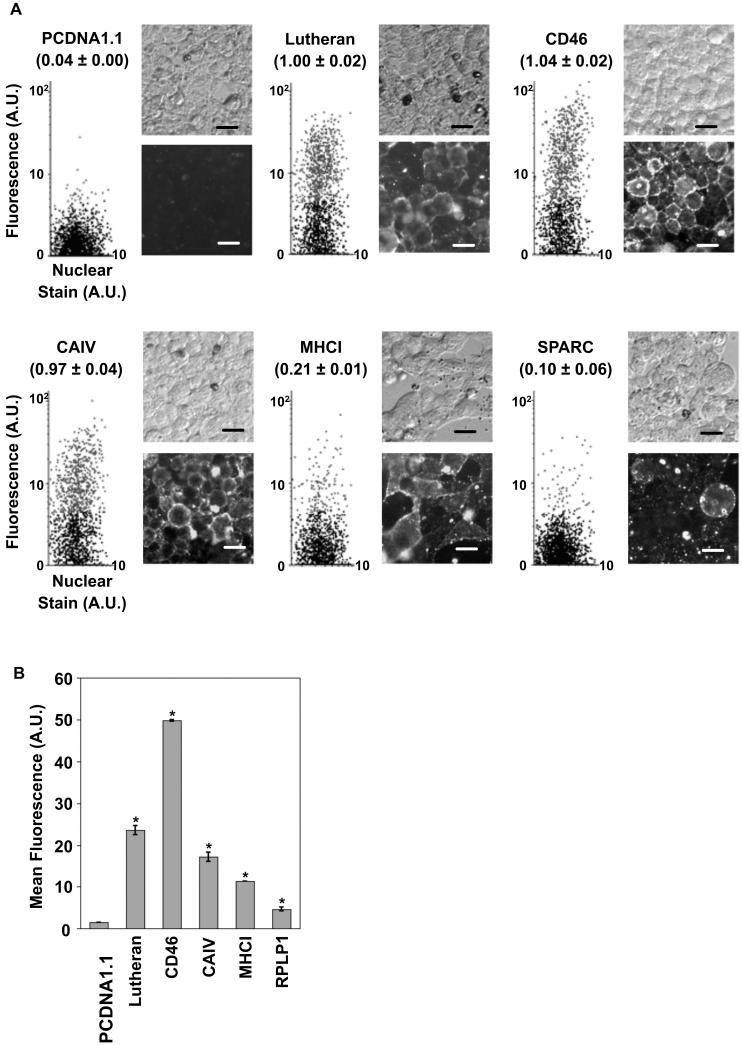
A total of 211 individual cDNA clones were recovered from the fourth round and of these, 30 were positively recognized by the BMSPA in flow cytometric analyses. Sequencing of the 30 clones yielded 15 different proteins as several clones were redundant. The identities of the 15 unique clones are listed in Table 1. The BMSPA immunoreactivity of HEK293 cells transfected with these individual cDNA clones was confirmed by both flow cytometry and immunocytochemistry, as shown in figure 6A for a few representative clones. As evident from figure 6A, there are significant differences in the fraction of transfected HEK293 cells that get immunolabeled for different membrane proteins, such as MHCI and SPARC as compared to Lutheran, CD46 or CAIV. This variation is likely the effect of several factors including levels of cell surface expression of the proteins and the titer of specific antibodies in the BMSPA. Importantly, whereas the previously described expression cloning approach [12] might have missed clones with low BMSPA immunoreactivity such as SPARC (Figure 6A), the flow cytometry based MEC approach developed here is sensitive enough to be able to consistently detect these clones. As seen in Table 1, which reports the in vivo transcript abundance levels for homologues of the identified proteins at the rat BBB [4], MEC has the power to recover clones that possess low abundance mRNA in vivo and hence correspond to low copy number cDNAs in the cDNA library. Despite the strong CMV promoter, the in vitro expression levels of the BBB membrane proteins in the host cells can also vary significantly as seen in Figure 6B, although they still remain within the detection limits of MEC.
Table 1.
BBB Membrane proteins identified using the MEC approach
| Symbol | Protein Name | Genbanka | Locationb | Transcript Abundancec | BBBd |
|---|---|---|---|---|---|
| CD46 | Membrane cofactor protein | NM_183080.1 | TM | 0 | [13] |
| CAIV | Carbonic anhydrase IV | NM_173897 | GPI | 4 | [22] |
| RACK1 | Receptor for activated C-kinase | BC102286 | MPA | 2 | [4] |
| LAMP2 | Lysosomal associated membrane protein 2 | BC102819 | TM | 9 | [4] |
| LU | Lutheran blood group (Auberger B antigen included) | NM_174741 | TM | 4 | [12] |
| SPARC | Secreted protein acidic cysteine rich / osteonectin | BT030548 | ECM | 295 | [4,5] |
| ITM2B | Integral membrane protein 2B | XM_587730 | TM | 29 | [23] |
| MHCI | Major histocompatibility complex class I, A | AY960156 | TM | 4 | [1, 5] |
| SCAMP1 | Secretory carrier membrane protein 1 | NM_001076054 | TM | 0 | |
| UCP2 | Mitochondrial uncoupling protein 2 | NM_001033611 | TM | 4 | [4] |
| SVIL | Supervillin | BC113265 | MPA | 0 | |
| RPLP1 | Ribosomal acidic protein large P1 | BC102695 | 116 | [4] | |
| ND4 | Mitochondrial NADH dehydrogenase subunit 4 | YP_209214 | TM | 108 | [4] |
| CTNNA1 | α-catenin related protein | XM_872743 | MPA | 3 | [4] |
| p85α | Phosphatidylinositol 3-kinase regulatory subunit 1 | NM_174575 | MPA | 0 |
Accession numbers from BLAST search against the nucleotide collection database (nr/nt) for Bos taurus derived sequences
TM-transmembrane, MPA-Membrane protein associated, ECM-Extracellular matrix, GPI-glycosylphosphatidylinositol associated
TA-transcript abundance reported as tags per 100,000 (from ref. [4], GSE3696 record [GEO; NCBI]). A transcript abundance of zero indicates that the sequence was not detected in the ref. [4] study
References indicating BBB localization.
Figure 6.
Analysis of individual membrane protein-encoding clones A. Representative images of HEK293 cells transfected with individual BBB membrane protein clones and immunolabeled with the BMSPA, including flow cytometric dot plots (left), phase contrast image (top) and the corresponding fluorescent image (bottom). Values in parentheses indicate the fraction of transfected cells normalized to the Lutheran transfected cells, along with SEM. Scale bar = 20 μm. B. Representative data for the mean fluorescence values, denoting cell surface immunolabeling intensity for selected BBB membrane protein clones. Error bars denote SEM for 2 measurements each; * = p<0.05 as compared to the mean fluorescence of PCDNA1.1 transfected cells, determined by a two-tailed Student's t-test.
Eight of the proteins listed in Table 1 contain at least one transmembrane domain while others, classified as membrane protein-associated, do not possess any transmembrane domains. Since the BMSPA was raised against intact plasma membrane sheets [15], recognition of both these classes of membrane proteins is not surprising. Finally, by design, the MEC approach is strongly biased towards the identification of membrane proteins that are truly BBB-resident by using a cDNA library derived from BBB mRNA and by using a BMSPA that was raised selectively against intact BBB membrane sheets and their associated proteins. In other words, both the protein and mRNA for the proteins listed in Table 1 are required to be present at the BBB for selection by the MEC process. Indeed, most of the membrane proteins identified in the current study have been reported to be present at the BBB by previous genomic or detailed protein characterization studies, as indicated in Table 1. For example, Carbonic anhydrase IV has been demonstrated to be a BBB-associated marker in rat and mouse brains [22]. Suppression subtractive hybridization studies conducted in rat and human models have shown the BBB localization of MHC I, SPARC and ITM2B transcripts [3, 23] and a more comprehensive SAGE-based genomic profiling of the rat BBB [4] identified RACK1, LAMP2, UCP2, RPLP1, ND4 and CTNNA1 as BBB-localized. The current study now confirms that the transcripts isolated in these genomic studies are indeed expressed as proteins at the BBB, given their reactivity with the BMSPA. Of the proteins listed in table 1, those that were not yet specifically identified as BBB-expressed either as gene or protein, include SCAMP1, SVIL and p85α.
4. DISCUSSION
In the current study, we developed the MEC approach to further analyze the membrane proteome of the BBB. Of the proteins identified in this study (Table 1), the Lutheran glycoprotein and CD46 were previously found to be selectively expressed at the bovine BBB in our precursor expression cloning studies [12, 13]. The Lutheran glycoprotein plays a role in basal cell adhesion [24] and is overexpressed in human brain tumor vessels [25], whereas CD46 is a complement regulator protein and has been shown to act as a receptor for the measles virus [26]. Another protein that serves a structural role is SVIL, which is involved in determining cell architecture at focal adhesions [27]. ITM2B and SPARC are both crucial in developing tissue. ITM2B is implicated in the differentiation of neuronal cells and is also connected to the hereditary familial British and Danish dementia neurodegenerative disorders [31]. The expression of ITM2B was found to be downregulated substantially in in vitro cultures of rat brain endothelial cells, potentially indicating its role in the maintenance of the BBB [23]. SPARC is an extracellular matrix-associated glycoprotein that functions in the maintenance of the extracellular matrix, angiogenesis and wound healing [28]. Due to its capability of dissociating focal adhesions in endothelial cells [28], this protein is likely an important factor in the regulation and remodeling of endothelial barriers in the brain and elsewhere in the body [29]. SPARC has also been reported to be overexpressed at the brain tumor vasculature by both quantitative transcript profiling and immunohistochemistry [30]. Interestingly, localization of this secreted protein was at the sites of perivascular cells [30]. This raises the point that while the proteins in Table 1 are of BBB origin for reasons described in the results section above, we cannot completely eliminate the possibility that certain proteins may be expressed by perivascular cells (pericytes or smooth muscle cells) that are, as a result of well-known technical limitations, part of BBB isolates used for cDNA and BMSPA preparation [1].
Apart from controlling the structural aspects of the BBB, some of the proteins identified in this study are essential for regulating other BBB functions. For example, carbonic anhydrase IV, a known BBB biomarker, may play a role in regulating the flux of CO2 and HCO3- across the BBB [22]. P85α, a ubiquitously expressed protein, serves as a regulatory subunit for phosphatidylinositol 3-kinase and is implicated in insulin resistance [32, 33]. RACK1 plays a role in several central nervous system activities including regulation of intracellular Ca2+ levels and neuronal excitation as well as determining sensitivity to ethanol [34]. Because of these diverse functions, RACK1 has been implicated in brain aging in addition to neurodegenerative disorders and epilepsy [34].
The clones identified by the BMSPA also included intracellular plasma membrane-associated and mitochondrial / endosomal membrane proteins. SCAMP1, ND4, UCP2, LAMP2 and RPLP1 are all examples of this class of proteins. Although these proteins primarily reside in other interior cell membranes, they are often also found at the plasma membrane surface which allowed isolation using the MEC approach. SCAMP1, a four pass transmembrane protein, is postulated to be involved in vesicle trafficking and is usually associated with trans-Golgi and endosomal membranes, though it has also been shown to be expressed on the plasma membrane of NRK cells [35]. ND4, a highly hydrophobic membrane protein with 12 predicted transmembrane helices, is a subunit of the mitochondrial NADH dehydrogenase complex I [36]. Mutations in ND4 are linked to Leber's hereditary optic neuropathy [37] and Leigh syndrome [38]. UCP2, another mitochondrial protein identified, dissipates the proton gradient across the inner mitochondrial membrane, reduces production of reactive oxygen species, simulates the biogenesis of new mitochondria and may directly influence synaptic plasticity and neuronal transmissions [39]. It has also been recently shown to be critical for Ca2+ uptake by mitochondria [40]. UCP2 prevents oxidative stress to cells thus acting as a neuroprotector in traumatic brain injury and ischemia and due to its strategic functions, may be involved in several neurodegenerative conditions [39]. LAMP2 is a highly glycosylated protein that localizes to lysosomal, endosomal and autophagic vacuole membranes [41] and is also expressed on the surface of several cell types including metastatic tumor cells [42]. The ribosomal protein RPLP1 identified by the BMSPA in this study has been previously reported to be expressed on the surface of rat mesangial cells and cell lines derived from rat brain astrocytes and mouse fibroblasts [43] and may play roles in the regulation of protein synthesis as well as in DNA transcription and repair [44].
The wide range of physical properties, function and abundance of the BBB membrane proteins identified in this study demonstrate the powerful attributes of the MEC approach. In addition, MEC yields full-length cDNAs that provide the user with flexibility to design future experiments for more detailed protein structure, function and localization studies. Although, the focus of this study was not the identification of membrane proteins specific to the BBB, the MEC approach developed here could also be used to identify only those membrane proteins that are selectively expressed at the BBB by first depleting the BMSPA against other vascular beds [12, 13, 14].
ACKNOWLEDGEMENTS
The authors wish to acknowledge Dr. William Pardridge for his generous gift of the bovine BBB membrane protein-specific polyclonal antiserum, the staff at the UW Paul P. Carbone Comprehensive Cancer Center Flow Cytometry Facility for their assistance and Johnson's Sausage Shoppe, Rio, WI for supplying bovine tissue. This project was funded by National Institutes of Health Grant NS052649.
List of abbreviations
- BBB
Blood-brain barrier
- BMSPA
BBB membrane protein-specific polyclonal antiserum
- CMV
Cytomegalovirus
- FACS
Fluorescence activated cell sorting
- MEC
Multiplex expression cloning
- SV40
Simian Virus 40
REFERENCES
- [1].Shusta EV. Blood-brain barrier genomics, proteomics, and new transporter discovery. NeuroRx. 2005;2:151–161. doi: 10.1602/neurorx.2.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Madden SL, Cook BP, Nacht M, Weber WD, et al. Vascular gene expression in nonneoplastic and malignant brain. Am. J. Pathol. 2004;165:601–608. doi: 10.1016/s0002-9440(10)63324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Calabria AR, Shusta EV. Blood-brain barrier genomics and proteomics: elucidating phenotype, identifying disease targets and enabling brain drug delivery. Drug Discov. Today. 2006;11:792–799. doi: 10.1016/j.drudis.2006.07.006. [DOI] [PubMed] [Google Scholar]
- [4].Enerson BE, Drewes LR. The rat blood-brain barrier transcriptome. J. Cereb. Blood Flow Metab. 2006;26:959–973. doi: 10.1038/sj.jcbfm.9600249. [DOI] [PubMed] [Google Scholar]
- [5].Pardridge WM. Blood-brain barrier genomics. Stroke. 2007;38:686–690. doi: 10.1161/01.STR.0000247887.61831.74. [DOI] [PubMed] [Google Scholar]
- [6].Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- [7].Schindler J, Lewandrowski U, Sickmann A, Friauf E, Nothwang HG. Proteomic analysis of brain plasma membranes isolated by affinity two-phase partitioning. Mol. Cell. Proteomics. 2006;5:390–400. doi: 10.1074/mcp.T500017-MCP200. [DOI] [PubMed] [Google Scholar]
- [8].Wang H, Qian WJ, Chin MH, Petyuk VA, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J. Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu L, Han DK. Overcoming the dynamic range problem in mass spectrometry-based shotgun proteomics. Expert Rev. Proteomics. 2006;3:611–619. doi: 10.1586/14789450.3.6.611. [DOI] [PubMed] [Google Scholar]
- [10].Cech NB, Enke CG. Relating electrospray ionization response to nonpolar character of small peptides. Anal. Chem. 2000;72:2717–2723. doi: 10.1021/ac9914869. [DOI] [PubMed] [Google Scholar]
- [11].Grant KJ, Wu CC. Advances in neuromembrane proteomics: efforts towards a comprehensive analysis of membrane proteins in the brain. Brief Funct. Genomic Proteomic. 2007;6:59–69. doi: 10.1093/bfgp/elm001. [DOI] [PubMed] [Google Scholar]
- [12].Shusta EV, Boado RJ, Pardridge WM. Vascular proteomics and subtractive antibody expression cloning. Mol. Cell. Proteomics. 2002;1:75–82. doi: 10.1074/mcp.t100008-mcp200. [DOI] [PubMed] [Google Scholar]
- [13].Shusta EV, Zhu C, Boado RJ, Pardridge WM. Subtractive expression cloning reveals high expression of CD46 at the blood-brain barrier. J. Neuropathol. Exp. Neurol. 2002;61:597–604. doi: 10.1093/jnen/61.7.597. [DOI] [PubMed] [Google Scholar]
- [14].Shusta EV, Li JY, Boado RJ, Pardridge WM. The Ro52/SS-A autoantigen has elevated expression at the brain microvasculature. Neuroreport. 2003;14:1861–1865. doi: 10.1097/00001756-200310060-00021. [DOI] [PubMed] [Google Scholar]
- [15].Pardridge WM, Yang J, Buciak JL, Boado RJ. Differential Expression of 53- and 45-kDa Brain Capillary-Specific Proteins by Brain Capillary Endothelium and Choroid Plexus in Vivo and by Brain Capillary Endothelium in Tissue Culture. Mol. Cell. Neurosci. 1990;1:20–28. doi: 10.1016/1044-7431(90)90038-6. [DOI] [PubMed] [Google Scholar]
- [16].Jordan M, Wurm F. Transfection of adherent and suspended cells by calcium phosphate. Methods. 2004;33:136–143. doi: 10.1016/j.ymeth.2003.11.011. [DOI] [PubMed] [Google Scholar]
- [17].Mellon P, Parker V, Gluzman Y, Maniatis T. Identification of DNA sequences required for transcription of the human alpha 1-globin gene in a new SV40 host-vector system. Cell. 1981;27:279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- [18].Chakrabarti S, Joffe S, Seidman MM. Recombination and deletion of sequences in shuttle vector plasmids in mammalian cells. Mol. Cell. Biol. 1985;5:2265–2271. doi: 10.1128/mcb.5.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- [20].Miller JH, Lebkowski JS, Greisen KS, Calos MP. Specificity of mutations induced in transfected DNA by mammalian cells. EMBO J. 1984;3:3117–3121. doi: 10.1002/j.1460-2075.1984.tb02267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lebkowski JS, DuBridge RB, Antell EA, Greisen KS, Calos MP. Transfected DNA is mutated in monkey, mouse, and human cells. Mol. Cell. Biol. 1984;4:1951–1960. doi: 10.1128/mcb.4.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ghandour MS, Langley OK, Zhu XL, Waheed A, Sly WS. Carbonic anhydrase IV on brain capillary endothelial cells: a marker associated with the blood-brain barrier. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6823–6827. doi: 10.1073/pnas.89.15.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Calabria AR, Shusta EV. A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. J. Cereb. Blood Flow Metab. 2008;28:135–148. doi: 10.1038/sj.jcbfm.9600518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Udani M, Zen Q, Cottman M, Leonard N, et al. Basal cell adhesion molecule/lutheran protein. The receptor critical for sickle cell adhesion to laminin. J. Clin. Invest. 1998;101:2550–2558. doi: 10.1172/JCI1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boado RJ, Li JY, Pardridge WM. Selective Lutheran glycoprotein gene expression at the blood-brain barrier in normal brain and in human brain tumors. J. Cereb. Blood Flow Metab. 2000;20:1096–1102. doi: 10.1097/00004647-200007000-00009. [DOI] [PubMed] [Google Scholar]
- [26].Seya T, Hirano A, Matsumoto M, Nomura M, Ueda S. Human membrane cofactor protein (MCP, CD46): multiple isoforms and functions. Int. J. Biochem. Cell Biol. 1999;31:1255–1260. doi: 10.1016/s1357-2725(99)00092-8. [DOI] [PubMed] [Google Scholar]
- [27].Wulfkuhle JD, Donina IE, Stark NH, Pope RK, et al. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J. Cell. Sci. 1999;112:2125–2136. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- [28].Bradshaw AD, Sage EH. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J. Clin. Invest. 2001;107:1049–1054. doi: 10.1172/JCI12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Goldblum SE, Ding X, Funk SE, Sage EH. SPARC (secreted protein acidic and rich in cysteine) regulates endothelial cell shape and barrier function. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3448–3452. doi: 10.1073/pnas.91.8.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pen A, Moreno MJ, Martin J, Stanimirovic DB. Molecular markers of extracellular matrix remodeling in glioblastoma vessels: microarray study of laser-captured glioblastoma vessels. Glia. 2007;55:559–572. doi: 10.1002/glia.20481. [DOI] [PubMed] [Google Scholar]
- [31].Vidal R, Delisle MB, Ghetti B. Neurodegeneration caused by proteins with an aberrant carboxyl-terminus. J. Neuropathol. Exp. Neurol. 2004;63:787–800. doi: 10.1093/jnen/63.8.787. [DOI] [PubMed] [Google Scholar]
- [32].Backer JM, Myers MG, Jr, Shoelson SE, Chin DJ, et al. Phosphatidylinositol 3'-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- [34].Sklan EH, Podoly E, Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog. Neurobiol. 2006;78:117–134. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- [35].Castle A, Castle D. Ubiquitously expressed secretory carrier membrane proteins (SCAMPs) 1-4 mark different pathways and exhibit limited constitutive trafficking to and from the cell surface. J. Cell. Sci. 2005;118:3769–3780. doi: 10.1242/jcs.02503. [DOI] [PubMed] [Google Scholar]
- [36].Bourges I, Ramus C, Mousson de Camaret B, Beugnot R, et al. Structural organization of mitochondrial human complex I: role of the ND4 and ND5 mitochondria-encoded subunits and interaction with prohibitin. Biochem. J. 2004;383:491–499. doi: 10.1042/BJ20040256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yen MY, Wang AG, Wei YH. Leber's hereditary optic neuropathy: a multifactorial disease. Prog. Retin. Eye Res. 2006;25:381–396. doi: 10.1016/j.preteyeres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- [38].Vanniarajan A, Rajshekher GP, Joshi MB, Reddy AG, et al. Novel mitochondrial mutation in the ND4 gene associated with Leigh syndrome. Acta Neurol. Scand. 2006;114:350–353. doi: 10.1111/j.1600-0404.2006.00673.x. [DOI] [PubMed] [Google Scholar]
- [39].Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nat. Rev. Neurosci. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- [40].Trenker M, Malli R, Fertschai I, Levak-Frank S, Graier WF. Uncoupling proteins 2 and 3 are fundamental for mitochondrial Ca2+ uniport. Nat. Cell Biol. 2007;9:445–452. doi: 10.1038/ncb1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–145. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- [42].Saitoh O, Wang WC, Lotan R, Fukuda M. Differential glycosylation and cell surface expression of lysosomal membrane glycoproteins in sublines of a human colon cancer exhibiting distinct metastatic potentials. J. Biol. Chem. 1992;267:5700–5711. [PubMed] [Google Scholar]
- [43].Sun KH, Liu WT, Tang SJ, Tsai CY, et al. The expression of acidic ribosomal phosphoproteins on the surface membrane of different tissues in autoimmune and normal mice which are the target molecules for anti-double-stranded DNA antibodies. Immunology. 1996;87:362–371. [PMC free article] [PubMed] [Google Scholar]
- [44].Tchorzewski M. The acidic ribosomal P proteins. Int. J. Biochem. Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]