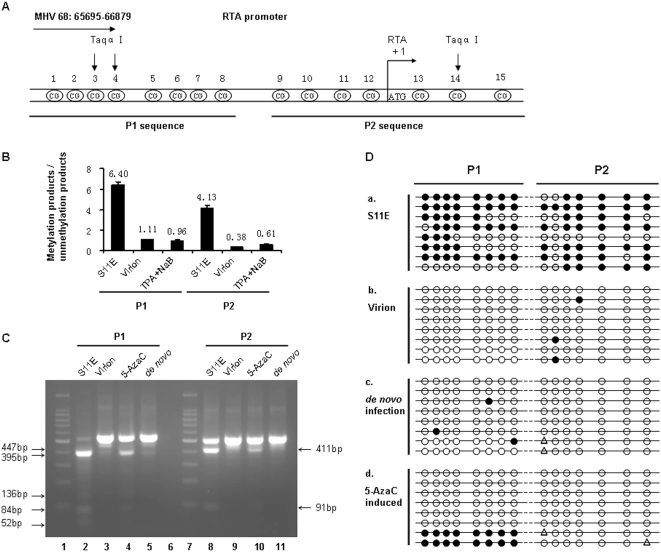
Figure 2. Methylation status at the MHV-68 RTA promoter.
(A) A schematic view of the RTA promoter on MHV-68 genome (NC_001826, n.t. 65695-66879). The 15 CpG sites shown were analyzed in this study, the promoter region divided into two parts for analysis, named P1 and P2. (B) Quantitative methylation-specific PCR (Q-MSP) analysis of the RTA promoter methylation status. Genomic DNAs were prepared from untreated S11E cells, MHV-68 virion or TPA (25 ng/ml) plus NaB (4 mM) treated S11E cells. Then Q-MSP was performed to detect methylation status of the RTA promoter P1 and P2 fragments, using methylation-specific (M) or unmethylation-specific (U) primers, respectively. Standard reactions were performed at the same time to quantify copy numbers, and the ratio of methylation-specific and unmethylation-specific products was calculated for each sample. Results represent the average from three experiments, with standard deviations shown. (C) Combined bisulfite restriction analysis (COBRA) of the MHV-68 RTA promoter methylation status. Genomic DNAs were prepared from untreated S11E cells, MHV-68 virion, 5-AzaC (10 µM, 36 hrs) induced S11E cells or de novo infected MHV-68 BHK-21 cells, bisulfite modified, and subsequently used for PCR amplification. The PCR products were digested by Taqα| for COBRA. The digested products for P1 fragments are 84 bp, 52 bp, 136 bp, 447 bp and 395 bp. The digested products for P2 fragments are 411 bp and 91 bp. (D) Bisulfite genomic sequencing (BGS) analysis of the 15 CpG sites at the RTA promoter. Genomic DNAs from untreated S11E cells, MHV-68 virion, de novo infected MHV-68 BHK-21 cells or 5-AzaC (10 µM, 24 hrs) induced S11E cells, bisulfite modified, and then used for PCR amplification, The PCR products were cloned into T-A vector. Each group included eight independent clones. Solid circles indicate methylated CpG, open circles indicate unmethylated CpG and triangle indicate ambiguous sequencing results.