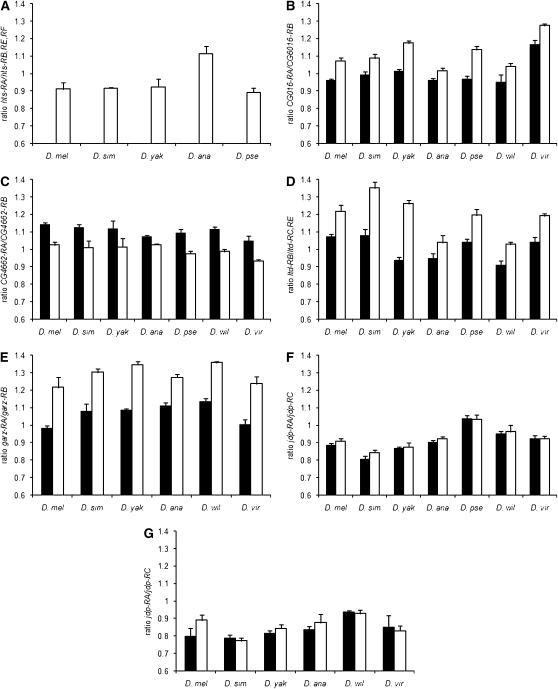
Figure 3.—
Sex-specific expression of alternative transcripts for six genes that showed significant probe-by-sex interaction in the microarray data (solid bars, males; open bars, females) among Drosophila species. The RT–PCR results are analyzed as the average ratio of CT values for the two exon junctions amplified for each gene (y-axis), and error bars show the standard deviation. D. mel, D. melanogaster; D. sim, D. simulans; D. yak, D. yakuba; D. ana, D. anannasae; D. pse, D. pseudoobscura; D. wil, D. willistoni; D. vir, D. virilis. (A) Ratio of hts-RA and hts-RB, -RE, and -RF for female whole bodies of D. melanogaster, D. simulans, D. yakuba, D. anannasae, and D. pseudoobscura. (B) Ratio of CG6016-RA and CG6016-RB male and female whole bodies for D. melanogaster, D. simulans, D. yakuba, D. anannasae, D. pseudoobscura, D. willistoni, and D. virilis. (C) Ratio of CG4662-RA and CG4662-RB male and female whole bodies for all seven species. (D) Ratio of ltd-RB and ltd-RC and -RE for male and female whole bodies for all seven species. (E) Ratio of garz-RA and garz-RB for male and female whole bodies of all seven species. (F) Ratio of jdp-RA and jdp-RC for male and female whole bodies tested for all seven species. (G) Ratio of jdp-RA and jdp-RC for male and female heads for D. melanogaster, D. simulans, D. yakuba, D. anannasae, D. willistoni, and D. virilis.