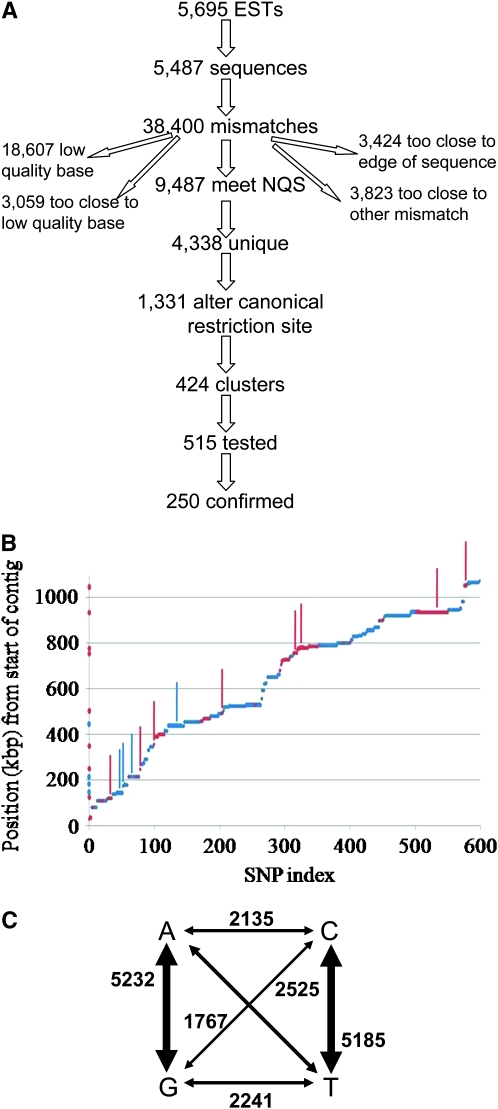
Figure 1.—
SNP discovery. (A) Of all sequenced MV ESTs, 5487 aligned consistently with the OR genome, and these contained 38,400 mismatches. Of these, 9487 met NQS criteria (see materials and methods); the remainder was excluded for the reasons shown. Because of redundancy in the library, these corresponded to 4338 unique putative SNPs (pSNPs), of which 1331 alter 1 of 10 common four-base restriction sites. These were divided over 424 clusters (see materials and methods), and at least one pSNP in each cluster was tested until 250 were unambiguously confirmed. (B) Distribution of 599 putative SNPs along contig 6. Position from the start of the contig (in kilobase pairs) is plotted as a function of pSNP index (rank order of the pSNP's physical position along the contig). Note the long horizontal stretches showing groups of closely linked and thus functionally redundant SNPs. Red dots denote pSNPs included in genomewide validation; blue dots denote other pSNPs; red bars (projected on y-axis for clarity) mark confirmed SNPs from genomewide validation; blue bars mark confirmed SNPs from regional validation. (C) Frequency of transition and transversion events (based on complete set of 17,394 putative SNPs).