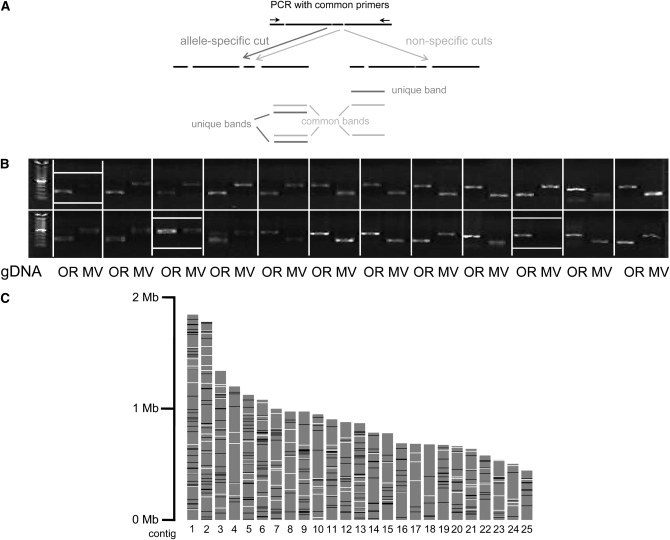
Figure 2.—
SNP validation. (A) Principle of cleaved amplified polymorphic sequence (CAPS) assay. A genomic region containing an allele-specific restriction site (dark gray, present in the left and absent in the right allele) and possibly common restriction sites (light gray) is amplified by PCR with a common pair of primers. Subsequent digestion produces several fragments, some of which are unique to one allele. These can be visualized by gel electrophoresis. (B) Representative gel for SNP validation. Digested PCR fragments obtained from Oak Ridge (OR) and Mauriceville (MV) are loaded side by side for 24 pSNPs. Common negative results include failure to efficiently amplify from MV gDNA and absence of the expected extra site in MV (boxed). (C) SNP coverage in the 25 largest contigs (shaded bars). Available EST sequence (used for genomewide validation) is denoted by open and validated CAPS markers by solid bars (resolution 5 kbp).