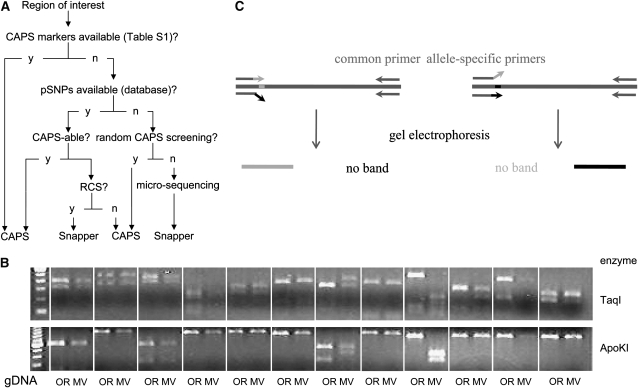
Figure 4.—
Development of local SNP markers. (A) General approach for efficient marker development. The first choice for markers in the region of interest consists of the set of validated CAPS markers described in supplemental Table S1. When these are exhausted but pSNPs are available, these can be developed as either CAPS or single nucleotide amplified polymorphism (SNAP) markers. Finally, additional polymorphisms can be found by random CAPS screening (RCS) or sequencing of randomly chosen PCR fragments. (B) Representative gel for random CAPS screening. Amplicons were prepared from 12 intergenic regions, using Oak Ridge (OR) or Mauriceville (MV) gDNA and digested with different enzymes. Note more irregular nature of digestion patterns compared to CAPS markers found by genomewide validation (Figure 2B). (C) Principle of SNAP assay. Two PCR reactions are carried out, using a common primer and either of two allele-specific primers. The latter are designed to be perfectly complementary to one allele but not the other, so that genomic DNA containing a specific allele is amplified only in the reaction employing its respective primer. Presence or absence of PCR products is visualized by gel electrophoresis.