Abstract
To counteract the problems associated with the purification of HIV envelopes, we developed a new purification method exploiting the high affinity of a peptide mimicking CD4 towards the viral glycoprotein. This miniCD4 was used as a ligand in affinity chromatography and allowed the separation in one step of HIV envelope monomer from cell supernatant and the capture of pre-purified trimer. This simple and robust method of purification yielded to active and intact HIV envelopes as proved by the binding of CCR5 HIV co-receptor, CD4 and a panel of well-characterized monoclonal antibodies. The immunogenicity of miniCD4-purified HIV envelope was further assessed in rats. The analysis of the humoral response indicated that elicited antibodies were able to recognize a broad range of HIV envelopes. Finally, this method based on a chemically synthesized peptide may represent a convenient and versatile tool for protein purification compatible far scale-up in both academic and pharmaceutical researches.
Keywords: HIV1, Monomer, Trimer, CD4 mimic, Affinity chromatography, AIDS vaccine
Introduction
Acquired immunodeficiency syndrome (AIDS) continues to be a major health problem throughout the world, with in 2007 approximately 33 million people infected with the human immunodeficiency virus type 1 (HIV-1), 2.5 million new infections, and 2.1 million people died of AIDS-related illnesses (UNAIDS/WHO, 2007). Therefore, there is an urgent need for an effective anti-HIV vaccine. The HIV-1 envelope glycoprotein (Env) is the target of broadly neutralizing antibodies and should be an important component of a successful HIV vaccine (Girard et al., 2006; Phogat and Wyatt, 2007). Env is formed by gp120 and gp41 that result of the cleavage of gp160 precursor (McCune et al., 1988). These two subunits are noncovalently associated in a trimeric structure (Wyatt and Sodroski, 1998). In order to mimic the native form of HIV-1 envelope, gp140 non-covalent trimer lacking gp41 trans-membrane domain has been engineered (Jeffs et al., 2004; Kieny et al., 1988; Sanders et al., 2002; Srivastava et al., 2002, 2003).
Recombinant HIV-1 glycoproteins are produced at high scale in both trimeric and monomeric forms for pre-clinical and clinical researches on HIV vaccine. Several studies have demonstrated that gp140 is particularly attractive as potent immunogen and that oligomers are more effective in inducing strong antibody responses to conformational epitopes compared to monomer (Barnett et al., 2008; Mascola et al., 1996; Srivastava et al,. 2003; Zhang et al., 2007). Indeed, the few monoclonal antibodies (mAbs) that potently neutralize HIV-1 all recognize epitopes exposed on the native Env complex (Burton and Parren, 2000, Fouts et al., 1998; Parren et al., 1998; Sattentau and Moore, 1995). In contrast, nonneutralizing mAbs that do not bind to the native complex have been found following vaccination with gp120 monomer (Burton and Parren, 2000, Parren et al., 1998). Therefore it is important to produce Env oligomers with intact epitopes and functional receptor binding sites for inclusion in a potential HIV-vaccine.
In order to address this point, we have developed a new method of purification of HIV-1 Env suitable for the capture of gp120 or gp140. There are a few pre-existing methods for the purification and the preparation of active Env. Two main approaches can be pinpointed. First, lectin is utilized in affinity chromatography to capture glycoproteins (Cefai et al., 1990; Jeffs et al., 2004; Jones et al., 1995, Srivastava et al., 2002, 2003). Unfortunately, this approach is unspecific to HIV-1 Env and requires several additional steps of purification such as ion exchange and size exclusion chromatography. Second, Env glycoproteins are purified with anti-gp120 or anti-gp140 mAbs (Jeffs et al., 2004; Kalyanaraman et al., 1990; Kwong et al, 1999). This approach presents several difficulties. Antibody-based methods are expensive and necessitate the coupling of anti-Env antibody to a solid support, which may reduce its affinity for Env. Moreover, there are few antibodies suitable for the purification of a large panel of envelope; and the half-life of antibody is relatively limited, particularly when used in acidic conditions.
Our objective was to define a new method of purification of HIV envelope, which was simple, robust, not expensive, suitable for envelope monomer or trimer and above all which preserved HIV envelope antigenicity and its ability to bind both CD4 and chemokine receptor. Therefore, we investigated the usefulness of a peptide mimicking the human receptor CD4 as a ligand in affinity chromatography. MiniCD4 peptides were initially obtained by the transfer of critical elements of CD4/gp120 interface onto a structurally compatible small scaffold (Martin et al., 2003; Vita et al., 1999). Their affinity towards gp120 and gp140 had been improved by combinatorial chemistry and directed amino-acid substitution based on the crystal structure of the complex gp120-miniCD4 (Huang et al., 2005; Stricher et al., 2005, 2008; Van Herrewege et al., 2008). In the affinity chromatography method presented here, we used CD4M48 an optimized version of miniCD4 that presents a sub-nanomolar affinity for SF162 isolate and a CD4-like affinity for clade B envelopes in neutralization assays (Stricher et al., 2008; Van Herrewege et al., 2008). The miniCD4 was grafted to a solid support and evaluated for its ability to purify gp120SF162 and gp120YU2 from mammalian cells supernatant. In parallel, the peptide was assessed for its capacity to capture gp140SF162 trimer. The yield of purification of each HIV-1 Env was measured and antigenic properties of miniCD4-purified Env were evaluated through the binding of CD4, CCR5 co-receptor and a panel of well-characterized neutralizing monoclonal antibodies. Finally, the immunogenicity of miniCD4-purified gp120YU2 was evaluated in a small scale animal trial.
Results
Purification of monomeric HIV envelope by miniCD4
A miniCD4-based affinity chromatography was designed to purify monomeric HIV-1 envelope from supernatant of cell over-expressing gp120SF162. 500 µl of cell supernatant (approximate concentration of gp120SF162: 150 mg/L) was incubated with 500 µl of miniCD4-beads. Bound proteins were then eluted in 1.5 ml of citric acid instantaneously neutralized by citrate buffer (see materials and methods).
The yield of miniCD4-affinity chromatography was evaluated by a CD4 binding assay through the measurement of the concentration of active gp120SF162 in cell supernatant and in the three fractions eluted from miniCD4-beads (Table 1). Typically, cell supernatant or eluted fractions (named E1–E3) were incubated in the presence of immobilized D7324 anti-gp120 mAb. Bound proteins were then detected after three subsequent incubations with (i) soluble CD4 (sCD4), (ii) L120.3 anti-CD4 mAb, (iii) a HRP labeled antibody directed against L120.3 mAb. Concentrations of gp120SF162 before and after miniCD4 capture were evaluated by comparison to several dilutions of gp120SF162 at known concentrations (gp120SF162 purified with the multi-step method). In the whole study, the multi-step method of purification of HIV envelope was use as a method of reference (see materials and methods). This method involving a capture and two cleaning steps had been shown to efficiently purify both HIV envelope monomer and trimer (Srivastava et al., 2002, 2003). We carried out 9 independent experiments and obtained 55.8 % as average yield of purification with the miniCD4 affinity chromatography method.
Table 1.
yield of purification of miniCD4 affinity chromatography
| Step | Volume (ml) | gp120SF162 (µg) | % of recovery | Purity (%) |
|---|---|---|---|---|
| Culture supernatant | 0.5 | 72.25 +/− 12.5 | ||
| MiniCD4 eluate | 1.5 | 40.3 +/− 4.12 | 55.8 | 60 |
Number of independent experiments: n= 9
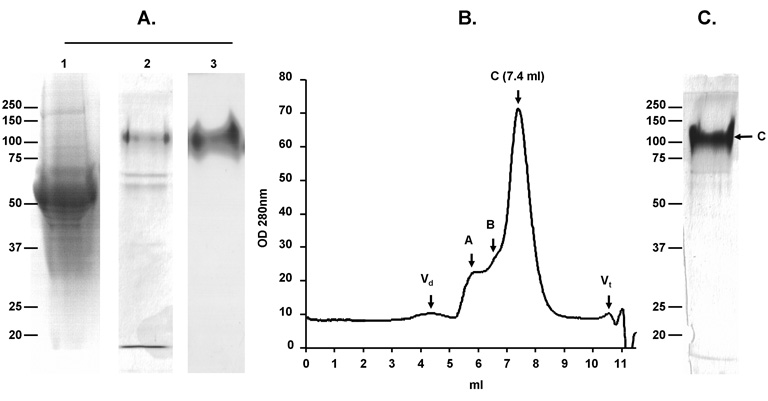
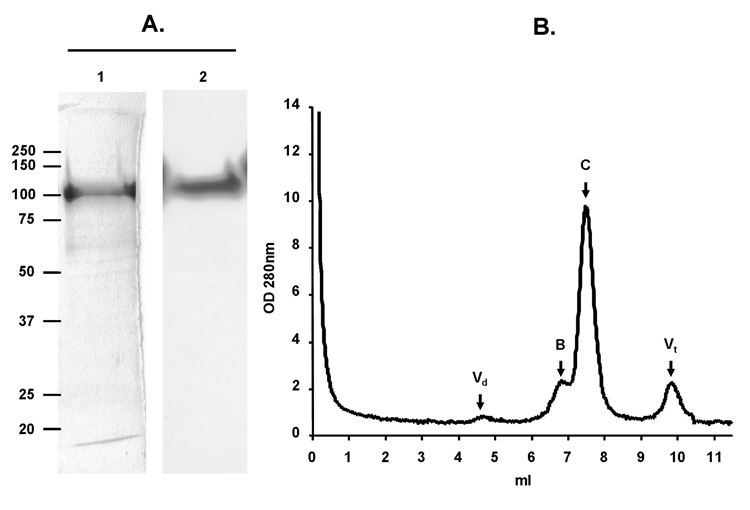
At the same time, captured proteins were analyzed by SDS-PAGE, immunoblot, and size exclusion chromatography (SEC) (Fig. 1). Purification of gp120SF162 was achieved with a degree of purity of approximately 60% as shown by SDS-PAGE followed by silver staining (Fig. 1A lane 2). The major contaminant was shown to be a serum protein, identified by N-terminal sequencing as bovine serum albumin precursor (Fig. 1A lane 2). The identity of HIV envelope was confirmed by immunoblot using anti-gp120 D7324 mAb (Fig. 1A lane 3). MiniCD4-purified gp120SF162 was further analyzed on a pre-calibrated Bio-Sil 250 column (Fig. 1B). Consistent with SDS-PAGE analysis, the elution profile shows that only a low amount of aggregate was eluted in death volume and peak A fractions. The elution volume of peak C fractions corresponds to protein with gp120 molecular weight (7.4 ml). Peak B fractions could be composed of gp120 aggregates or by non-envelope proteins found in cell supernatant, which possess a size of 300 KDa (6.5 ml). The fractions constituting peak C were pooled and analyzed by SDS-PAGE (Fig. 1C). Silver staining revealed a purity of gp120SF162 estimated at > 90 % by densitometric scanning.
Fig. 1. Purification of gp120SF162 by the miniCD4 method.
Analysis of miniCD4-purified gp120SF162 by SDS-PAGE (A). Lanes l and 2 silver staining: lane 1—flowthrough after miniCD4 beads incubation; lane 2—pool of miniCD4 elutions. Lane 3 immunoblot: identification of gp120SF162 among the eluted protein by anti-gp120 D7324 mAb. Size exclusion-HPLC profile of miniCD4-purified gp120SF162 (B). Fractions composing peak C were pooled and analyzed by SDS-PAGE followed by a silver staining (C).
Binding of miniCD4-purified gp120SF162 to CCR5 receptor
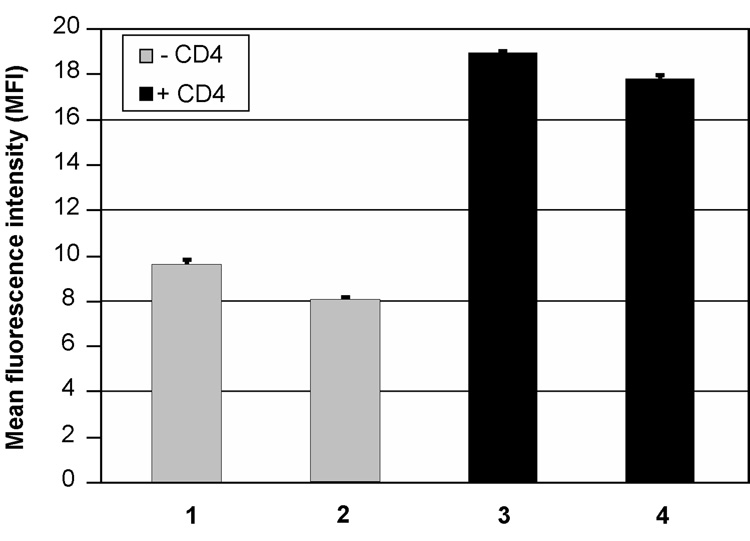
The interaction of CD4 with gp120 allows the subsequent binding of gp120 to a chemokine receptor through an important conformational change. In order to evaluate the activity of gp120SF162 purified from miniCD4 affinity chromatography, HIV envelope was pre-incubated or not with soluble CD4 (sCD4) and then assessed for its binding activity towards CCR5 chemokine receptor. Addition of sCD4 increased the binding of gp120SF162 purified by miniCD4 affinity chromatography to CCR5 receptor (Fig. 2, comparison of lane 1 and lane 3), leading to a binding activity comparable to gp120 purified by the reference method and associated with sCD4 (Fig. 2, lane 3 compared to lane 4).
Fig. 2. Binding of miniCD4-purified gp120SF162 to CCR5 chemokine receptor.
Over-expressing CCR5 cells were incubated with 100 ng of gp120SF162 without (lanes 1 and 2) or with 36 ng of sCD4 (lanes 3, 4). Bound envelope glycoprotein was detected with D7324 anti-gp120 mAb. Cells were analyzed for envelope binding by FACS flow cytometry. Lane 1—miniCD4-purified gp120SF162; lane 2—gp120SF162 purified by the reference method; lane 3—miniCD4-purified gp120SF162+ sCD4; lane 4—gp120SF162 purified by the reference method + sCD4.
Immunochemical characterization of miniCD4-purified gp120SF162
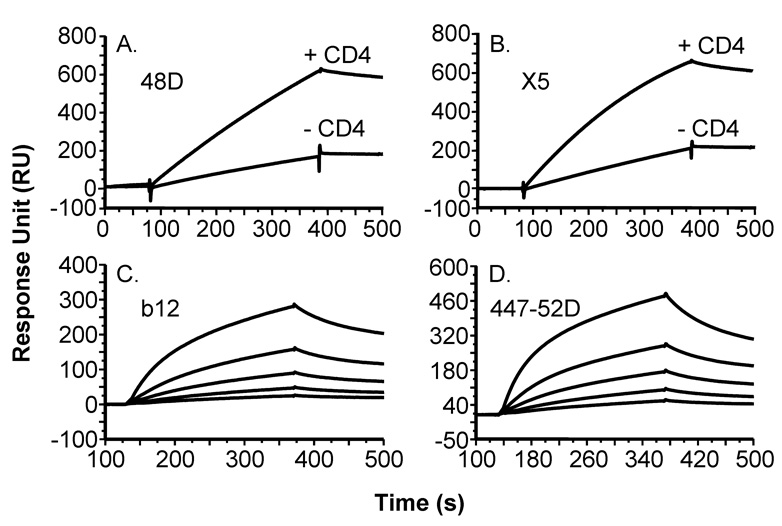
The structural integrity of gp120SF162 purified with the miniCD4 affinity chromatography method was further analyzed by surface immunoprobing with a panel of mAbs of defined epitope specificities, using Surface Plasmon Resonance (SPR) and capture ELISA. Upon binding to sCD4, miniCD4-purified gp120SF162 underwent conformational changes as reflected by an increase binding of X5 and 48D CD4-induced mAbs (CD4i Abs; Fig. 3A and B). MiniCD4-purified gp120SF162 was also recognized by 44752D anti-V3 loop and b12 anti-CD4 binding site mAbs (Fig. 3C and D).
Fig. 3. Binding sensorgrams of miniCD4-purified gp120SF162 to anti-gp120 mAbs.
The binding of miniCD4-purified gp120SF162 on 48D (A) or X5 (B) antibody, in the presence or not of 100 nM sCD4 (soluble CD4) was analyzed by surface plasmon resonance (SPR). Binding sensorgrams of miniCD4-purified gp120SF162 diluted at 50 nM, 25 nM, 12.5 nM, 6.25 nM and 3.125 nM to b12 (C) or 44752D (D) mAbs.
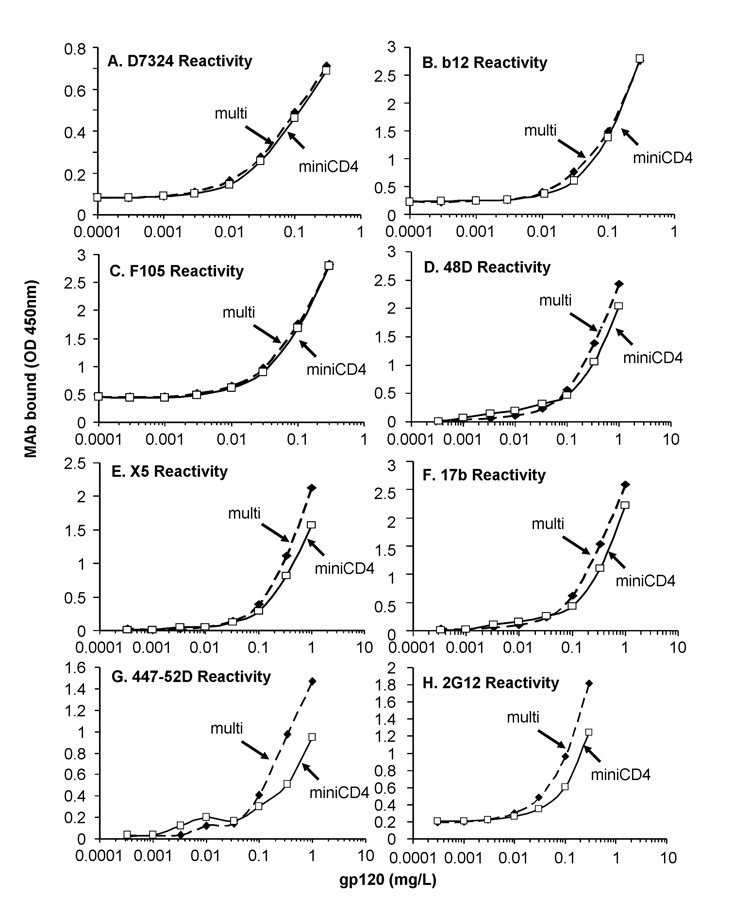
The miniCD4 affinity chromatography was further compared to the reference method. HIV envelope gp120SF162 purified by the two methods were analyzed for their ability to bind a panel of well-defined mAbs. First, the concentration of HIV envelopes was normalized through ELISA assays using D7324 mAb (Fig. 4A). Then, gp120SF162 envelopes purified by miniCD4 (named miniCD4) or by the reference method (named multi) were assessed by using mAbs directed against CD4i epitopes (X5, 48D and 17b), CD4 binding site (b12, F105), carbohydrate moieties (2G12) and V3 loop (44752D). Despite the moderate degree of purity of gp120SF162 purified in one step by miniCD4 (Fig. 1A), a similar antibody-binding was observed for both gp120SF162 glycoproteins (Fig. 4B–H), suggesting that miniCD4 allows the separation of HIV-1 envelope with intact epitopes.
Fig. 4. Comparison of the binding to a panel of well-characterized mAbs of miniCD4-purified gp120SF162 and gp120SF162 purified with a multi-step method.
Various concentrations of miniCD4-purified gp120SF162 (open square) and gp120SF162 purified with a multi-step method (reference method, filled square) were pre-incubated in the presence or not of an equimolar concentration of soluble CD4 (addition of sCD4 for panels D, E and F) on immobilized lectin (Concanavalin A) and incubated with several mAbs i.e. D7324 (A); b12 (B); F105 (C); 48D (D); X5 (E); 17b (F); 44752D (G); 2G12 (H). Anti-gp120 mAbs binding was finally detected with peroxidase-labeled secondary antibodies. Figure 4 shows the representative antibody-binding signal of 4 independent experiments.
Legends: “multi”: gp120SF162 purified with the multi-step procedure, “miniCD4”: gp120SF162 purified by miniCD4 affinity chromatography.
Purification of trimeric gp140 by miniCD4
MiniCD4 affinity chromatography was further assessed for its ability to capture gp140 trimer. 50 µg of gp140SF162ΔV2 purified by the method of reference were diluted in 500 µl of PBS and incubated 1 hour at room temperature with an equal volume of miniCD4-beads. Then, unbound and bound proteins from three independent experiments were collected in a final volume of 500 µl each. All the fractions were further analyzed by spectrometry at 280 nm and by SPR (evaluation of gp140 binding to 48D CD4i mAb). A percentage of 92.4 ± 2.2 % of gp140 captured by miniCD4 was measured by spectrometry, a comparable yield of purification was observed by SPR.
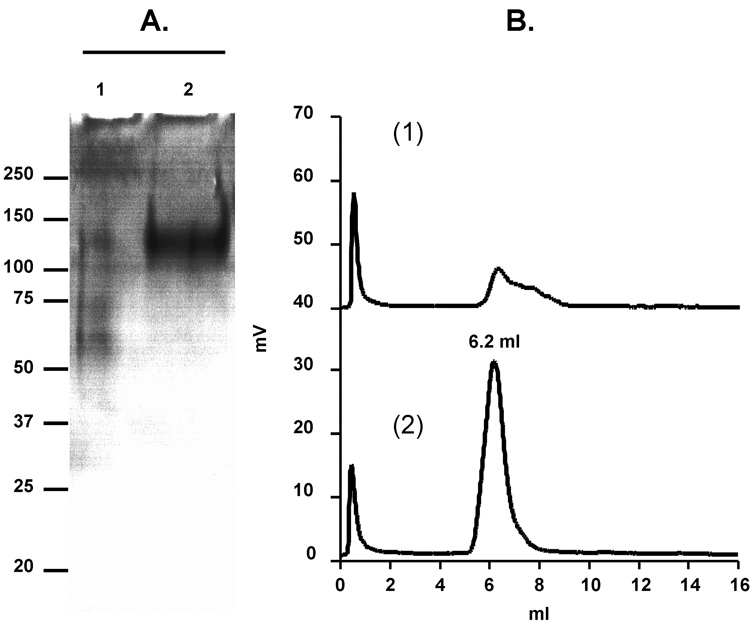
MiniCD4-purified gp140SF162ΔV2 was further analyzed by SDS-PAGE, immunoblot and SEC (Fig. 5). MiniCD4 affinity chromatography allowed the elimination of contaminating proteins of lower molecular mass (Fig. 5A, comparison of lane 1 and lane 2). MiniCD4 affinity chromatography appeared efficient to reinforce the grade of purity of pre-purified gp140, contaminants proteins of 50–70 KDa size should be composed of Bovine Serum Albumin, as previously shown for gp120 (Fig. 5A, line 1). Consistent with the yield of purification described above, a very low proportion of protein with gp140 molecular weight was found in unbound fractions (Fig. 5A, lane 1). As expected, miniCD4-purified gp140SF162ΔV2 migrated with an apparent molecular mass of 140 kDa in reducing and denaturing conditions due to the presence of gp41 ectodomain downstream gp120 core protein.
Fig. 5. Purification of gp140SF162ΔV2 by the miniCD4 method.
SF162 envelope trimer purified by the multi-step method was applied to miniCD4 beads. Analysis of miniCD4-purified gp140SF162ΔV2 by SDS-PAGE (A: lane 1—flow through after miniCD4 beads incubation; lane 2—pool of miniCD4 elutions). Size exclusion-HPLC profile of miniCD4-purified o-gp140SF162ΔV2 (B; panel 1—flow through after miniCD4 beads incubation; panel 2—pool of miniCD4 elutions).
MiniCD4-purified gp140SF162ΔV2 was further analyzed over pre-calibrated size exclusion HPLC column (Bio-Sil SEC-250, Fig. 5B, panel 2). Based on the volume of elution of gp120 (7.4 ml), gp140 (6.2 ml) and standards (670 kDa = 5.52 ml; 158 kDa = 7.12 ml; 44 kDa = 7.79 ml; 17 kDa = 9.28 ml and 1.35 kDa = 10.55 ml), the calculated molecular masses of gp120SF162 and gp140SF162ΔV2 are 132.3 and 418.3 kDa, respectively. The molecular mass of purified gp140SF162ΔV2 is the expected molecular mass for a trimer (3 × 140 kDa [monomer] = 420 kDa [trimer]), while gp120 one is slightly higher. Consistent with the previous experiments, the SEC profile of unbound fractions indicates the presence of a low proportion of gp140 and contaminating proteins of lower molecular weight (Fig. 5B, panel 1).
Immunochemical characterization of miniCD4-purified gp140SF162ΔV2
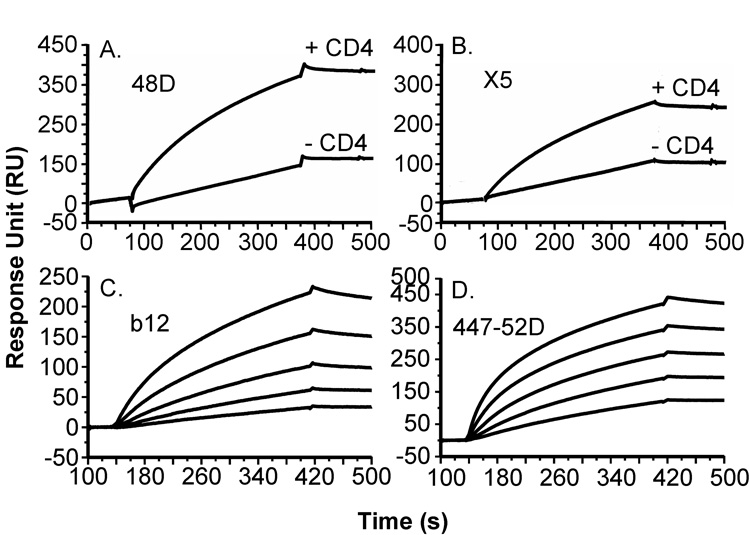
The structural integrity of gp140SF162ΔV2 purified with miniCD4 affinity chromatography was further evaluated by SPR using the panel of antibodies described for gp120SF162. MiniCD4-purified gp140 showed an increased affinity for both X5 and 48D CD4i mAbs in the presence of sCD4 (Fig. 6A and B), and was recognized by b12 and 44752D mAbs (Fig. 6C and D, respectively). Interestingly, gp140SF162ΔV2 koff is reduced for all the antibodies tested compared to gp120SF162 one (see Fig. 3 and Fig. 6), suggesting an increased avidity for gp140 consistent with the trimeric conformation observed by SEC (Fig. 5B).
Fig. 6. Binding sensorgrams of miniCD4-purified gp140SF162ΔV2 to anti-gp120 mAbs.
The binding of miniCD4-purified gp140SF162ΔV2 on 48D (A) or X5 (B) antibody, in the presence or not of 100 nM sCD4 was analyzed by SPR. Binding sensorgrams of miniCD4-purified gp140SF162ΔV2 diluted at 50 nM, 25 nM, 12.5 nM, 6.25 nM and 3.125 nM to b12 (C) or 44752D (D) mAbs.
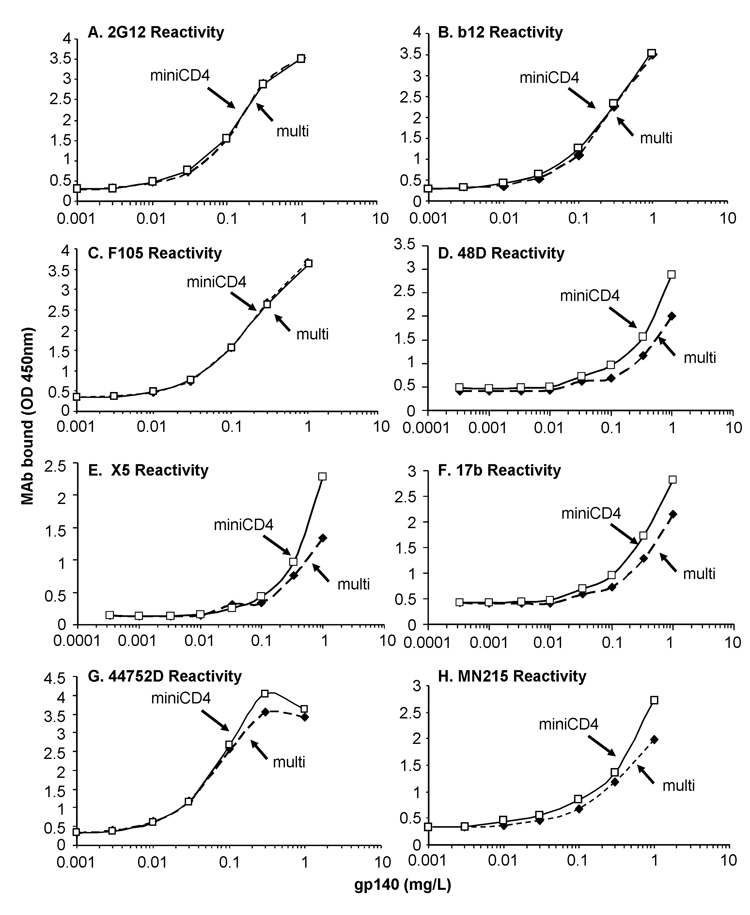
As described above for gp120SF162, gp140ΔV2SF162 purified by miniCD4 was finally compared to gp140ΔV2SF162 purified by the reference method exclusively. Both gp140ΔV2SF162 concentrations were normalized in ELISA by using 2G12 mAb (Fig. 7A). Afterwards, the two trimers were analyzed for their ability to bind mAbs described above (X5, 48D, 17b, b12, F105, 44752D and MN215 mAbs). We found that SF162 trimer antigenicity was perfectly preserved by the miniCD4 method of purification (Fig. 7B–H). Taken together, these data underline the usefulness of miniCD4 chromatographic method for the purification of functional and intact Env trimer.
Fig. 7. Immunochemical characterization gp140SF162ΔV2 purified with the multi-step method and submitted (open square) or not (filled square) to miniCD4 capture.
Various concentrations of gp140SF162ΔV2 purified with the multi-step method and submitted (open square) or not (filled square) to miniCD4 capture were pre-incubated on immobilized lectin (Concanavalin A) and incubated with a panel of mAbs i.e. 2G12 (A); b12 (B); F105 (C); 48D (D); X5 (E); 17b (F); 44752D (G); MN215 (H). Anti-gp120 mAbs binding was finally detected with peroxidase-labeled secondary antibodies. For X5, 48D and 17b binding gp140SF162ΔV2 was pre-incubated in the presence of an equimolar concentration of sCD4. Figure 7 shows the representative antibody-binding signal of 4 independent experiments.
Legends: “multi”: gp140ΔV2SF162 purified by the multi-step procedure, “miniCD4”: gp140ΔV2SF162 purified by miniCD4 affinity chromatography.
Immunogenicity of miniCD4-purified envelopes
MiniCD4 affinity chromatography was used to purify 1 mg of gp120YU2 from 1 liter of supernatant from cells over-expressing the viral envelope (approximate concentration of gp120YU2 in cell supernatant: 2 mg/L). In one step, gp120YU2 with a high grade of purity was obtained, as shown by SDS-PAGE, immunoblot and SEC analysis (Fig. 8A and B). Afterwards, three Wistar rats were immunized with miniCD4-purified gp120YU2 or miniCD4 peptide (used as control) using an intra-peritoneal boosting regimen. After the second and the third intramuscular boosts with gp120YU2, an important titer of antibody directed against the HIV envelope was observed. Sera from the three rats immunized by either gp120YU2 or miniCD4 were pooled and IgG were purified by affinity chromatography using G protein (Amersham). Then antibodies elicited by the immunogens were evaluated for their ability to recognize a panel of HIV-1 envelope monomers and trimers from diverse genetic sub-types (clades B, C and F). Table 2 presents the dilutions of antibody resulting in half-maximum binding to HIV-1 envelopes. MiniCD4-purified gp120YU2 elicited a high titre of antibody directed against envelopes monomers from B and C clades. However, elicited antibodies were less efficient in recognizing gp140 trimers from B, C and F clades; suggesting a certain proportion of antibody directed against envelope domains which are inaccessible on oligomer surface. This small scale immunogenicity study indicates that miniCD4-purified gp120 is suitable for vaccine candidate preparation. Furthermore, taken that miniCD4-purified envelopes possess an intact CD4 binding site, this method of purification may be particularly appropriate for the preparation of antigen aiming to induce antibodies directed against the CD4 binding site.
Fig. 8. Purification of gp120YU2 by the miniCD4 method.
Analysis of miniCD4-purified gp120YU2 by SDS-PAGE (A: lane 1, silver staining: pool of proteins eluted from miniCD4-beads; lane 2, immunoblot: identification of gp120YU2 among the eluted protein by anti-gp120 D7324 mAb) and size exclusion-HPLC profile of miniCD4-purified gp120YU2 (B).
Table 2.
Dilution of purified antibody resulting in half-maximum binding to several Envelope glycoproteins
| Antisera | Antigens |
|
|---|---|---|
| miniCD4-purified gp120YU2 | miniCD4 | |
| gp120CN54 (C) | 3000 | <100 |
| gp120HxB2 (B) | 1000 | <100 |
| gp120LAI (B) | 600 | <100 |
| gp120SF162 (B) | 1550 | <100 |
| gp120SF2 (B) | 1400 | <100 |
| gp120W61D (B) | 2000 | <100 |
| gp120YU2 (B) | 2000 | <100 |
| gp140Br29 (F) | 250 | <100 |
| gp140Bx08 (B) | 450 | <100 |
| gp140ZM96 (C) | 225 | <100 |
Discussion
To address the many problems associated with the purification of HIV envelopes and particularly the separation of intact HIV envelope trimer, we exploited the affinity of miniCD4 for gp120 and gp140 to develop an innovative method suitable for the purification of highly active and immunogenic HIV envelope.
HIV-1 envelope glycoprotein represents a major target for the development of AIDS vaccines. Initial trials of HIV-1 Env-based vaccines showed that soluble recombinant envelope glycoproteins were well-tolerated and elicited neutralizing antibodies to the homologous vaccine strain, but not to heterologous primary virus isolates (Graham and Wright, 1995; Jeffs et al., 2004; Mascola et al., 1996). Furthermore, these vaccines elicited sterilizing immunity against homologous virus and a restricted panel of heterologous virus in non-human primate models (Berman et al., 1990; Girard et al., 1995, 1996; Stott et al., 1998). Those observations had led to numerous envelope-based vaccines evaluated at both preclinical and clinical stages (Graham, 2002; Graham and Mascola 2005; Gallo 2005; Girard et al., 2006; Phogat and Wyatt, 2007). Nevertheless, the achievement of these different vaccine candidates requires important quantities of recombinant envelope monomer and/or trimer with intact epitopes and functional CD4-binding sites. Therefore, it is necessary to have a simple and reproducible method that lets the purification of active Env in good yields.
Pre-existing methods are mainly based on lectin capture or anti-gp120, anti-gp140 mAb affinity chromatography. These methods may require the conjunction of several steps of purification such as ion exchange and SEC and in most cases are specific to one HIV-1 envelope. To avoid a multi-step procedure and the use of anti-Env antibody, we used a miniCD4 peptide presenting a high affinity for gp120 and gp140 (Martin et al., 2003; Stricher et al., 2005, 2008, Van Herrewege et al., 2008; Vita et al., 1999). We showed that miniCD4 allowed the separation in one step of either gp120SF162 or gp120YU2 from cell supernatant. MiniCD4-purified gp120SF162 exhibited a high affinity for CD4, CCR5 and a panel of well-characterized mAbs directed against the CD4 binding site, CD4i epitopes and V3 loop. The affinity of miniCD4-purified gp120SF162 towards those ligands was comparable to the one of gp120SF162 purified with a multi-step procedure (Srivastava et al., 2002, 2003). Taken together, these data suggest that the use of acidic conditions for the elution of gp120SF162 does not affect the antigenic properties of the envelope glycoprotein. Finally, we performed an immunogenicity study indicating that miniCD4 method of purification is suitable for vaccine preparation, as miniCD4-purified gp120YU2 elicited anti-envelope antibodies directed against a broad range of genetic sub-types.
Taken the important need of envelope oligomers presenting intact epitopes for vaccine application, miniCD4 affinity chromatography was also evaluated for its ability to capture Env trimer. In order to achieve better immunogenecity with Env based-vaccines, attention has focused on Env oligomer rather than monomer. The association of envelope subunits in a trimeric conformation on HIV-1 surface shields potential epitopes. Thus, antibodies induced by gp120 monomer react more strongly to monomer as compared to oligomer (Burton and Parren, 2000, Broder et al., 1994; Parren et al., 1998; VanCott et al., 1995). Moreover, it has been well documented that gp120 monomers are limited in their ability to neutralize primary isolates (Mascola et al., 1996; Moore et al., 1995; VanCott et al., 1995; Wrin and Nunberg, 1994). Taken together, those observations had led to the engineering of several gp140 trimers and their evaluation as vaccine candidate (Barnett et al., 2008; Jeffs et al., 2004; Srivastava et al., 2002, 2003, 2008; Zhang et al., 2007). There are relatively few published studies that describe the purification of recombinant gp140 from eukaryotic expression systems. The non-covalent association of the different subunits forming gp140 trimer renders difficult the purification of envelope oligomers with classical method such as lectin capture and antibody-affinity chromatography. Indeed, Jeffs and others had noted the propensity of recombinant gp140/gp160 to form high molecular weight aggregates, particularly when lectin affinity chromatography is used (Binley et al., 2000; Jeffs et al., 2004; Rhodes et al., 1994). In this study, we found that miniCD4 was able to efficiently capture gp140ΔV2SF162 trimer and that miniCD4-purified trimer presented intact epitope as shown by CD4 and mAbs binding. We also showed that the grade of purity of pre-purified gp140ΔV2SF162 can be slightly increased after the miniCD4 purification step. Indeed, we showed that miniCD4-eluted gp140 was separated from lower molecular mass contaminants. Moreover, we did not observe aggregate formation following the miniCD4 affinity purification. However, to address precisely this point, miniCD4 purification should be repeated on cell supernatant of cell over-expressing gp140 rather than pre-purified oligomer.
The advantages of the use of miniCD4 in a purification method are many fold. First, miniCD4 presents a high stability compared to monoclonal antibodies. Indeed, miniCD4 is not affected by pH ranging from 2 to 9, variation of temperature from 20° to 90 °C and presents an important resistance to proteolytic degradation. Thus, miniCD4 beads can be stored at 4°C for a long period and reused several times without significant loss in their ability to capture Env. Second, because of its well-defined 3D structure and its reduced size (27 amino acids), miniCD4 can be easily and rapidly produced by chemical synthesis and labeled with affinity tag such as biotin moiety. In addition, as miniCD4 derives from a natural mini-protein is refolding is particularly easy (see material and method part). Third, in a recent study, we and others showed that CD4M48 and, more particularly, its derivative M48-U1 were able to neutralize a broad range of primary isolates (Van Herrewege et al., 2008). Those data emphasized the high affinity of miniCD4 for oligomeric envelopes belonging to a broad spectrum of HIV-1 primary strains. Thus, CD4M48 and M48-U1 could be advantageously used to purify subtype C gp140, as well as a large panel of Env from different clades. Fourth, another advantage inherent to miniCD4-purification is the selection of protein with accessible and well-folded CD4-binding site. This region is recognized by a broadly neutralizing antibody, b12 mAb (Binley et al., 2004, Ruprecht et al., 2003) which has been shown to present protective effects in non-human primate challenged with SHIV (simian/human immunodeficiency virus) (Parren et al., 1995, 2001; Ruprecht et al., 2003; Veazey et al., 2003). A recent study has shown that b12 mAb binding site has considerable overlap with both CD4 and miniCD4 ones (Huang et al., 2005; Zhou et al., 2007), suggesting that the selection of envelopes with intact miniCD4 binding site might be particularly attractive in vaccine preparation. Finally, the use of miniCD4 in a purification method represents a new example of applications of mini-protein engineered by the transfer of active site onto small-size scaffolds. Thus, biologically active mini-proteins may become not only useful tools in biology and medical research, but also a convenient step for protein purification in academic and pharmaceutical researches.
Materials and methods
All Fmoc (fluoren-9-ylmethoxycarbonyl)-protected amino acids were from Nova Biochem (via VWR international, Fontenay-sous-Bois, France). Fmoc PAL-PEG-PS resin was from Applied Biosystems (via Applera Corporation, Norwalk, CT, U.S.A.). All the other reagents and solvents used in the synthesis and purification of CD4M48 were from Fulka (St Quentin Fallavier, France) or SDS (Solvants Documentation Sytheses, Peypin, France). PBS and Biotin and unspecified chemical products were from Sigma Chemical Co. (St Louis, MO). The matrix used to bind the biotinylated miniCD4 was a Streptavidin Sepharose™ high performance from GE Healthcare Ltd (Uppsala, Sweden), the labeled miniCD4 was grafted to the beads at 1 µM final concentration.
Peptide synthesis
The miniCD4 CD4M48 was chemically synthesized on an Applied Biosystems Synthesizer (model 433) by solid-phase method using fluorenylmethyloxycarbonyl-protected amino acids and N-hydroxybenzotriazole/dicyclohexylcarbodiimide coupling strategy (Drakopoulou et al., 1996). The three disulfide bonds were formed with the peptides dissolved at 0.1mg/ml in 50 mM phosphate buffer (pH 7.8) in the presence of sequential addition of 5 mM reduced gluthatione followed by 0.5mM oxidized glutathione. Synthetic peptide was purified by reverse-phase HPLC, and its identity was verified by amino acid analysis and electrospray mass spectrometry. The biotin moiety was specifically introduced at Lys11 by using N-α-Fmoc-Nε-1-(ivDde-3-methylbutyl)-L-Lysine (where ivDde is 4,4-dimethyl-2,6-dioxocyclohex-1-ylidine) during peptide synthesis and subsequent coupling of Fmoc-8-amino-3,6-dioxaoctanoic acid (linker arm) and biotinamidohexanoic acid N-hydroxysuccinimide ester, after ivDde was removed by four treatments of 3 min with 2% hydrazine.
CD4M48 amino acid sequence is the following:
TpaNLHFCQLRC (K-Biot) SLGLLGRCAdPTFCACV
Lysine amino-acid was derivatized with a Biotin (K- Biot) moiety; dP was for (D)-proline and Tpa for thiopropionyl.
Envelope glycoproteins preparation
The CHO cell line DG-44 was used for generation of stable cell lines expressing either gp120SF162 (monomer) or o-gp140SF162ΔV2 (trimer with a partial deletion in the second variable loop V2) (Srivastava et al., 2002, 2003). The gene cassette used for the derivation of the stable cell lines contained the protein-encoding region of the envelope protein fused in frame to the human tissue plasminogen activator (t-PA) signal sequence. The signal peptide of t-PA has been previously described to increase the secretion of recombinant protein in the cell supernatant (Chapman et al., 1991).
Concerning gp120YU2, the HIV-1 envelope glycoprotein was produced in Spodotera frugiperda cells infected with Autographa californica baculovirus AcSLP10 (Mechulam et al., 2005). As previously described for the expression of HXB2, the natural signal peptide sequence of gp120YU2 was replaced by a new signal sequence isolated from the ecdysteroid glycosyltransferase (EGT) gene of AcSLP10 baculovirus in order to increase the secretion of the glycoprotein in cell supernatant (Misse et al., 1998).
MiniCD4 affinity chromatography
Cell supernatant from cells over-expressing gp120SF162 was concentrated 20-fold through a 100-kDa-pore-size membrane filter and stored at −80°C in the presence of 1 mM EDTA and 1 mM EGTA. Prior the purification process, the culture medium was filtrated over a 0.45 µm filter, and protease inhibitors were added. Afterward, 500 µl of culture medium were applied to 500 µl of miniCD4 beads (CD4M48 final concentration being approximately 1 µM) and incubated with gentle rocking overnight at 4°C. After 5 washes with 500 µl of PBS, proteins bound to the miniCD4 beads were recovered in 3 fractions of 250 µl of 25 mM Citric acid pH 3.6 each. Finally, eluted fractions were neutralized with 250 µl of 400 mM Citrate pH 6.8.
For scale-up growth, 1 liter of cell supernatant from cell infected with baculovirus producing gp120YU2 was divided in 200 ml fractions. Each fraction was then incubated with 10 ml of miniCD4 beads over night in rolling bottles at 4°C. As it was the case for the small scale analysis, the miniCD4 concentration in the mix was about 1 µM. Finally, bound material was eluted in 2.5 ml of acidic solution and neutralized with 2.5 ml of citrate solution as described above.
Multi-step method of purification used as control
HIV-1 envelope glycoprotein purified with the miniCD4 affinity chromatography was compared to Env obtained via another method of purification (Srivastava et al., 2002; 2003). Briefly, this multi-step procedure of purification is composed as followed, CHO cell supernatant was loaded onto a DEAE (GE Healthcare Ltd) column equilibrated with buffer (20 mM Tris, 100 mM NaCl, pH 8.0). Under these conditions, gp120SF162 and o-gp140SF162ΔV2 did not bind to the column, but contaminating proteins were retained on the column. The DEAE flowthrough was adjusted to 10 mM Phosphate and pH 6.8 and loaded on to a ceramic hydroxyapatite (CHAP, Biorad, Hercules, CA) column equilibrated with buffer (10 mM Na2HPO4, 100 mM NaCl, pH 6.8). Envs were recovered in the flowthrough, and the pH was adjusted to 7.4 with 2 M Tris (pH 8.8) and loaded on to a protein-A-agarose column to remove immunoglobulin contamination. The protein A-agarose flowthrough was loaded onto a Galanthus Nivalis-agarose (GNA) (Vector Laboratories, Burlingame, CA) column equilibrated with 20 mM Tris-100 mM NaCl (pH 7.4). Bound Envs were eluted with 500 mM methyl mannose pyranoside (Sigma Chemical Co.) in equilibrated buffer. All the fractions containing the HIV-1 envelope glycoproteins were pooled and fractioned on Superose-6 and Superdex-200 (GE Healthcare Ltd) tandem columns equilibrated with 10 mM Na-Citrate and 500 mM NaCl. Fractions containing Envs were pooled, concentrated using a Stir cell (Millipore, Inc., Bedford, MA), and stored at −80° until used. HIV-1 envelope glycoproteins purified with this approach were used through this study as control to quantify gp120SF162 and gp120YU2 obtained with the miniCD4 affinity chromatography. Envs obtained with this multi-step purification procedure were also termed ‘pre-purified Env’ and used to characterize the miniCD4-based method of purification.
Immunoblot analysis
Proteins were fractionated by SDS-PAGE and electro-blotted on PVDF membrane. Transferred proteins were detected by binding of anti-gp120 D7324 mAb (Aalto Bio Reagents, Dublin, Ireland) followed by a horseradish peroxidase conjugated anti-IgG sheep Ab. Immuno-reactive protein bands were detected by autoradiography (Kodak, Sigma) using an enhanced chemiluminescence assay (ECL, Amersham Biosciences, Piscataway, NJ).
CD4 binding assay by ELISA
The capability of purified envelope protein to bind CD4, was determined by using a sandwich ELISA assay (Stricher et al., 2005). Briefly, the 96-wells plates (Maxisorb; Nunc, Rochester, NY) were coated overnight at 4°C with D7324 mAb (50 ng per well); wells were saturated with PBS/5 % BSA buffer, wash three times, then different dilutions of purified gp120SF162 or gp120YU2 were added, followed by an incubation with 62 pg of sCD4 (Progenic, Tarrytown, NY). For detection, anti-CD4 mAb L120.3 (Centralized facilities for AIDS reagents, National Institute for Biological Standards and control (NIBSC), South Mimms, Potters Bar, Herts., U.K.) was added, followed by an incubation with goat-antimouse peroxidase-conjugated antibody (Jackson Immunoresearch, West Grove, PA) and substrate (3,3’,5,5’-tetramethylbenzidine; Sigma). The OD was measured at 450 nm and the concentration of purified gp120 was determined by comparison with standard curves derived from a known concentration of recombinant gp120SF162 purified with the multi-step method described above.
Immunological characterization of miniCD4-purified gp120
The binding of well-characterized HIV-Env specific mAbs was performed by a capture ELISA. Typically, the 96-wells plates were coated overnight at 4°C with lectin (concanavalin A, Sigma, 500 ng per well); wells were saturated with PBS/5 % BSA buffer, wash three times, and different dilutions of purified gp120SF162 were added. The capture proteins were revealed by incubation with 50 ng of anti-Env mAbs (Centralized facilities for AIDS reagents, NIBSC) 1 hour at room temperature. Several monoclonal antibodies were tested, directed against the CD4-binding site (b12 and F105), CD4-induced epitopes (17b, X5 and 48D), V3 loop (MN215 and 44752D), the carbohydrates moieties (2G12) and the C-terminal part of gp120 (D7324). For CD4-induced epitopes, gp120SF162 was pre-incubated with an equimolar concentration of miniCD4 before the incubation with immobilized-lectin. To finish, plates were washed, and specifically bound antibodies were detected using peroxidase-conjugated antibody as described earlier.
Size exclusion chromatography (SEC)
Purified gp120SF162, gp120YU2 and o-gp140 SF162ΔV2 were separated over pre-calibrated size exclusion HPLC column (Bio-Sil SEC-250, Biorad) by using ÄKTA purifier apparatus (Amersham Bioscience). Peak elution was monitored either at 280 nm or through Tryptophan fluorescence (λ excitation= 280 nm and λ emission= 340 nm, values expressed in mV: mvolts). Tryptophan fluorescence was measured with a spectrofluorometer FL-detector L-2480 (VWR international). Eluted proteins were calibrated by comparison with the SEC profile of molecular weight standards of 670, 158, 44, 17 and 1.35 kDa (Biorad).
Surface plasmon resonance (SPR) biosensor analysis
Experiments were conducted at 25° C with 20 µl/min flow rate in HBS (10mM HEPES-buffer saline, 3 mM EDTA, 0.05% Biacore surfactant, pH 7.4) with a Biacore 3000 instrument (GE Healthcare, Biacore AB, Uppsala, Sweden). All the antibodies tested were immobilized at ~1,500 RU by the amine coupling kit (NHS/EDC) provided by the manufacturer. We used the following panel of anti-HIV-1 Env mAbs: D7324, X5, 48D, b12 and 44752D. All the sensorgrams were corrected by subtracting the signal from reference flow cell.
Cell surface CCR5 chemokine receptor binding assays
Adherent CHO-K1 cells expressing CCR5 (Samson et al., 1996) were used to analyze envelope conformational change by flow cytometry FACS (Becton Dickinson, San Jose, CA). Typically, 100 ng of gp120SF162 or gp120YU2 (120 kDa) were pre-incubated or not with 36 ng of sCD4 (44kDa) or 2.5 ng of miniCD4 (3kDa) in 200 µl of PBS and then added to 50 µl of HAMF12-FCS 10% medium containing 2.105 CCR5 cells. After 1 h of incubation at room temperature, cells were washed 3 times with PBS/BSA 5 % (pH 7.4). Envelope binding to CCR5 co-receptor was further detected by a phycoerythrin-tagged antibody directed against D7324 antibody.
Immunogenicity studies
Young Wistar rats were immunized by intra-peritoneal path (i.p.) with 50 µg of miniCD4-purified gp120YU2 or 5 µg of miniCD4 CD4M48 emulsified in an equal volume of Alum adjuvant on day 0. Booster inoculations were given i.p. on day 28, 56 and 84. 2 milliliters of serum were collected at day 42, 70 and 98. Antibody titers were assayed using a sandwich ELISA as follows: plates were coated with Concanavalin A (500 ng per well) over night at 4°C, saturated with PBS/BSA 5%, pretreated 1h with gp120YU2 and then treated 1 h with collected sera. Bound antibodies were revealed with goat-anti-rat peroxidase-conjugated antibody.
Acknowledgements
We thank Drs. Sarah Bregant, Pascal Kessler and Andrej Galat for helpful discussion and assistance. This work was supported by grant “HIV Vaccine research and design, HIVRAD” PAR-00-093 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, supervised by Novartis vaccines and diagnostics, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnett SW, Srivastava IK, Kan E, Zhou F, Goodsell A, Cristillo AD, Ferrai MG, Weiss DE, Letvin NL, Montefiori D, Pal R, Vajdy M. Protection of macaques against vaginal SHIV challenge by systemic or mucosal and systemic vaccinations with HIV-envelope. AIDS. 2008;22(3):339–348. doi: 10.1097/QAD.0b013e3282f3ca57. [DOI] [PubMed] [Google Scholar]
- Berman PW, Gregory TJ, Riddle L, Nakamura GR, Champe MA, Porter JP, Wurm FM, Hershberg RD, Cobb EK, Eichberg JW. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature. 1990;345(6276):622–625. doi: 10.1038/345622a0. [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, Earl PL, Long D, Abedon ST, Moss B, Doms RW. Antigenic implications of human immunodeficiency virus type 1 envelope quaternary structure: oligomer-specific and -sensitive monoclonal antibodies. Proc. Natl. Acad. Sci. U. S. A. 1994;91(24):11699–11703. doi: 10.1073/pnas.91.24.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Parren PW. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat. Med. 2000;6(2):123–125. doi: 10.1038/72200. [DOI] [PubMed] [Google Scholar]
- Cefai D, Debre P, Kaczorek M, Idziorek T, Autran B, Bismuth G. Human immunodeficiency virus-1 glycoproteins gp120 and gp160 specifically inhibit the CD3/T cell-antigen receptor phosphoinositide transduction pathway. J. Clin. Invest. 1990;86(6):2117–2124. doi: 10.1172/JCI114950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman BS, Thayer RM, Vincent KA, Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19(14):3979–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakopoulou E, Zinn-Justin S, Guenneugues M, Gilquin B, Menez A, Vita C. Changing the structural context of a functional beta-hairpin. Synthesis and characterization of a chimera containing the curaremimetic loop of a snake toxin in the scorpion alpha/beta scaffold. J. Biol. Chem. 1996;271(20):11979–11987. doi: 10.1074/jbc.271.20.11979. [DOI] [PubMed] [Google Scholar]
- Fouts TR, Trkola A, Fung MS, Moore JP. Interactions of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate: relationship to neutralization. AIDS Res. Hum. Retrovir. 1998;14(7):591–597. doi: 10.1089/aid.1998.14.591. [DOI] [PubMed] [Google Scholar]
- Gallo RC. The end or the beginning of the drive to an HIV-preventive vaccine: a view from over 20 years. Lancet. 2005;366(9500):1894–1898. doi: 10.1016/S0140-6736(05)67395-3. [DOI] [PubMed] [Google Scholar]
- Girard M, Meignier B, Barré-Sinoussi F, Kieny MP, Matthews T, Muchmore E, Nara PL, Wei Q, Rimsky L, Weinhold K, et al. Vaccine-induced protection of chimpanzees against infection by a heterologous human immunodeficiency virus type 1. J. Virol. 1995;69(10):6239–6248. doi: 10.1128/jvi.69.10.6239-6248.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard M, Yue L, Barré-Sinoussi F, van der Ryst E, Meignier B, Muchmore E, Fultz PN. Failure of a human immunodeficiency virus type 1 (HIV-1) subtype B-derived vaccine to prevent infection of chimpanzees by an HIV-1 subtype E strain. J. Virol. 1996;70(11):8229–8233. doi: 10.1128/jvi.70.11.8229-8233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard MP, Osmanov SK, Kieny MP. A review of vaccine research and development: the human immunodeficiency virus (HIV) Vaccine. 2006;24(19):4062–4081. doi: 10.1016/j.vaccine.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Graham BS, wright PF. Candidate AIDS vaccines. N. Engl. J. Med. 1995;333(20):1331–1339. doi: 10.1056/NEJM199511163332007. [DOI] [PubMed] [Google Scholar]
- Graham BS. Clinical trials of HIV vaccines. Annu. Rev. Med. 2002;53:207–221. doi: 10.1146/annurev.med.53.082901.104035. [DOI] [PubMed] [Google Scholar]
- Graham BS, Mascola JR. Lessons from failure--preparing for future HIV-1 vaccine efficacy trials. J. Infect. Dis. 2005;191(5):647–649. doi: 10.1086/428406. [DOI] [PubMed] [Google Scholar]
- Huang CC, Stricher F, Martin L, Decker JM, Majeed S, Barthe P, Hendrickson WA, Robinson J, Roumestand C, Sodroski J, Wyatt R, Shaw GM, Vita C, Kwong PD. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13(5):755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Jeffs SA, Goriup S, Kebble B, Crane D, Bolgiano B, Sattentau Q, Jones S, Holmes H. Expression and characterization of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine. 2004;22(8):1032–1046. doi: 10.1016/j.vaccine.2003.08.042. [DOI] [PubMed] [Google Scholar]
- Joint United Nations programme on HIV/.AIDS (UNAIDS)/World Health Organization (WHO) Genewa Switzerland: AIDS epidemic update. 2007 December; 2007.
- Jones DH, McBride BW, Roff MA, Farrar GH. Efficient purification and rigorous characterisation of a recombinant gp120 for HIV vaccine studies. Vaccine. 1995;13(11):991–999. doi: 10.1016/0264-410x(95)00019-w. [DOI] [PubMed] [Google Scholar]
- Kalyanaraman VS, Rodriguez V, Veronese F, Rahman R, Lusso P, DeVico AL, Copeland T, Oroszlan S, Gallo RC, Sarngadharan MG. Characterization of the secreted native gp120 and gp160 of the human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 1990;6(3):371–380. doi: 10.1089/aid.1990.6.371. [DOI] [PubMed] [Google Scholar]
- Kieny MP, Lathe R, Rivière Y, Dott K, Schmitt D, Girard M, Montagnier L, Lecocq J. Improved antigenicity of the HIV env protein by cleavage site removal. Protein Eng. 1988;2(3):219–225. doi: 10.1093/protein/2.3.219. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Desjardins E, Robinson J, Culp JS, Hellmig BD, Sweet RW, Sodroski J, Hendrickson WA. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1) J. Biol. Chem. 1999;274(7):4115–4123. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- Martin L, Stricher F, Misse D, Sironi F, Pugniere M, Barthe P, Prado-Gotor R, Freulon I, Magne X, Roumestand C, Menez A, Lusso P, Veas F, Vita C. Rational design of a CD4 mimic that inhibits HIV-1 entry and exposes cryptic neutralization epitopes. Nat. Biotechnol. 2003;21(1):71–76. doi: 10.1038/nbt768. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, Keefer MC, McElrath MJ, Walker MC, Wagner KF, McNeil JG, McCutchan FE, Burke DS The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 1996;173(2):340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53(1):55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- Mechulam A, Cerutti M, Pugniere M, Misse D, Gajardo J, Roquet F, Robinson J, Veas F. Highly conserved beta16/beta17 beta-hairpin structure in human immunodeficiency virus type 1 YU2 gp120 is critical for CCR5 binding. J. Mol. Med. 2005;83(7):542–552. doi: 10.1007/s00109-005-0673-1. [DOI] [PubMed] [Google Scholar]
- Misse D, Cerutti M, Schmidt I, Jansen A, Devauchelle G, Jansen F, Veas F. Dissociation of the CD4 and CXCR4 binding properties of human immunodeficiency virus type 1 gp120 by deletion of the first putative alpha-helical conserved structure. J. Virol. 1998;72(9):7280–7288. doi: 10.1128/jvi.72.9.7280-7288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, 3rd, Burton DR, Ho DD. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 1995;69(1):101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Ditzel HJ, Gulizia RJ, Binley JM, Barbas CF, 3rd, Burton DR, Mosier DE. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS. 1995;9(6):F1–F6. doi: 10.1097/00002030-199506000-00001. [DOI] [PubMed] [Google Scholar]
- Parren PW, Mondor I, Naniche D, Ditzel HJ, Klasse PJ, Burton DR, Sattentau QJ. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 1998;72(5):3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, Moore JP, Burton DR. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 2001;75(17):8340–8347. doi: 10.1128/JVI.75.17.8340-8347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phogat S, Wyatt R. Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr. Pharm. Des. 2007;13(2):213–227. doi: 10.2174/138161207779313632. [DOI] [PubMed] [Google Scholar]
- Rhodes AD, Spitali M, Hutchinson G, Rud EW, Stephens PE. Expression, characterization and purification of simian immunodeficiency virus soluble, oligomerized gp160 from mammalian cells. J. Gen. Virol. 1994;75(Pt 1):207–213. doi: 10.1099/0022-1317-75-1-207. [DOI] [PubMed] [Google Scholar]
- Ruprecht RM, Ferrantelli F, Kitabwalla M, Xu W, McClure HM. Antibody protection: passive immunization of neonates against oral AIDS virus challenge. Vaccine. 2003;21(24):3370–3373. doi: 10.1016/s0264-410x(03)00335-9. [DOI] [PubMed] [Google Scholar]
- Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35(11):3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002;76(17):8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 1995;182(1):185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Legg H, Kan E, Fong A, Coates SR, Leung L, Wininger M, Donnelly JJ, Ulmer JB, Barnett SW. Purification and characterization of oligomeric envelope glycoprotein from a primary R5 subtype B human immunodeficiency virus. J. Virol. 2002;76(6):2835–2847. doi: 10.1128/JVI.76.6.2835-2847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Stamatatos L, Kan E, Vajdy M, Lian Y, Hilt S, Martin L, Vita C, Zhu P, Roux KH, Vojtech LC, Montefiori D, Donnelly J, Ulmer JB, Barnett SW. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 2003;77(20):11244–11259. doi: 10.1128/JVI.77.20.11244-11259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava IK, Kan E, Sun Y, Sharma VA, Cisto J, Burke B, Lian Y, Hilt S, Biron Z, Hartog K, Stamatatos L, Cheng RH, Ulmer JB, Barnett SW. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2008;372(2):273–290. doi: 10.1016/j.virol.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Stott EJ, Almond N, Kent K, Walker B, Hull R, Rose J, Silvera P, Sangster R, Corcoran T, Lines J, Silvera K, Luciw P, Murphy-Corb M, Momin P, Bruck C. Evaluation of a candidate human immunodeficiency virus type 1 (HIV-1) vaccine in macaques: effect of vaccination with HIV-1 gp120 on subsequent challenge with heterologous simian immunodeficiency virus-HIV-1 chimeric virus. J. Gen. Virol. 1998;79(Pt 3):423–432. doi: 10.1099/0022-1317-79-3-423. [DOI] [PubMed] [Google Scholar]
- Stricher F, Martin L, Barthe P, Pogenberg V, Mechulam A, Menez A, Roumestand C, Veas F, Royer C, Vita C. A high-throughput fluorescence polarization assay specific to the CD4 binding site of HIV-1 glycoproteins based on a fluorescein-labelled CD4 mimic. Biochem. J. 2005;390(Pt 1):29–39. doi: 10.1042/BJ20041953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricher F, Huang CC, Descours A, Duquesnoy S, Combes O, Decker JM, Do Kwon Y, Lusso P, Shaw GM, Vita C, Kwong PD, Martin L. Combinatorial Optimization of a CD4-Mimetic Miniprotein and Cocrystal Structures with HIV-1 gp120 Envelope Glycoprotein. J. Mol. Biol. 2008 doi: 10.1016/j.jmb.2008.06.069. In press; Ref: YJMBI 60586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Herrewege Y, Morellato L, Descours A, Aerts L, Michiels J, Heyndrickx L, Martin L, Vanham G. CD4 mimetic miniproteins: a new class of potent anti-HIV compounds with promising activity as microbicides. J. Antimicrob. Chemother. 2008;61(4):818–826. doi: 10.1093/jac/dkn042. [DOI] [PubMed] [Google Scholar]
- VanCott TC, Veit SC, Kalyanaraman V, Earl P, Birx DL. Characterization of a soluble, oligomeric HIV-1 gp160 protein as a potential immunogen. J. Immunol. Methods. 1995;183(1):103–117. doi: 10.1016/0022-1759(95)00038-c. [DOI] [PubMed] [Google Scholar]
- Veazey RS, Shattock RJ, Pope M, Kirijan JC, Jones J, Hu Q, Ketas T, Marx PA, Klasse PJ, Burton DR, Moore JP. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 2003;9(3):343–346. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
- Vita C, Drakopoulou E, Vizzavona J, Rochette S, Martin L, Menez A, Roumestand C, Yang YS, Ylisastigui L, Benjouad A, Gluckman JC. Rational engineering of a mini-protein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 1999;96(23):13091–13096. doi: 10.1073/pnas.96.23.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrin T, Nunberg JH. HIV-1MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS. 1994;8(11):1622–1623. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Zhang PF, Cham F, Dong M, Choudhary A, Bouma P, Zhang Z, Shao Y, Feng YR, Wang L, Mathy N, Voss G, Broder CC, Quinnan GV., Jr Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc. Natl. Acad. Sci. U. S. A. 2007;104(24):10193–10198. doi: 10.1073/pnas.0608635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445(7129):732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]