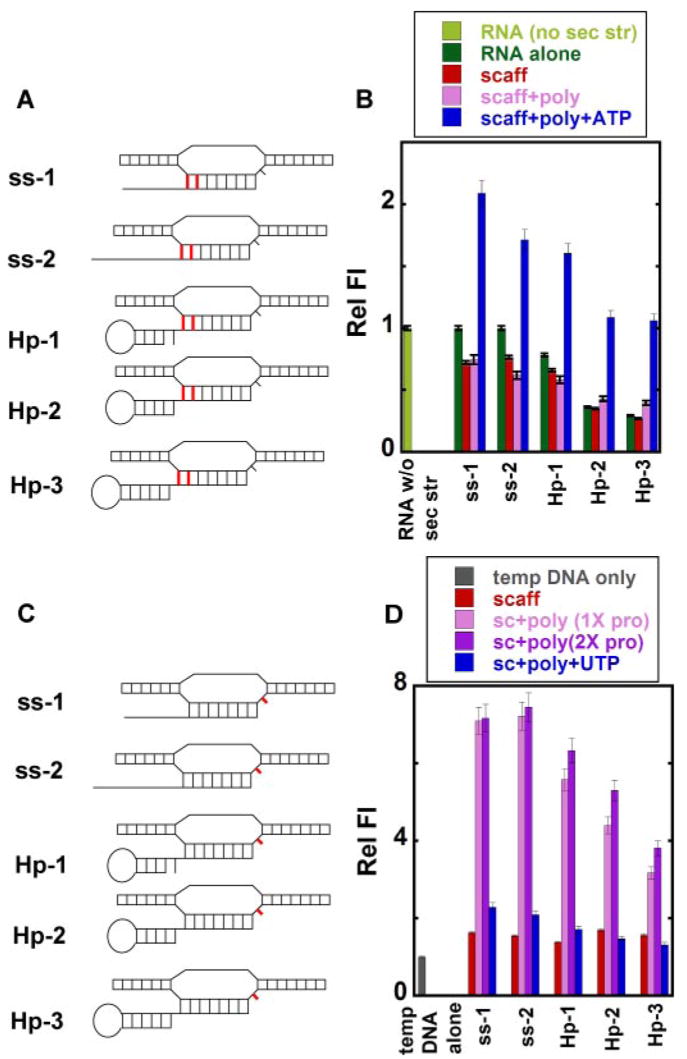
FIGURE 6. Local base conformations at the upstream end of the RNA-DNA hybrid and at the active site in free and RNAP-complexed nucleic acid scaffolds.
A and C, scaffolds showing the positions of 2-AP base(s) (in red) in the RNA and in the template DNA. B and D, relative fluorescence intensities. Light green, control RNA without secondary structure (sec str); dark green, RNA alone with the 2-AP bases in the RNA strand (A); gray, single-stranded template DNA with the 2-AP base in the template DNA (C); red, RNA-DNA scaffold; pink, scaffold-polymerase complex at equimolar concentrations; purple, scaffold-polymerase complex with twice the concentration of RNAP; blue, scaffold-RNAP complex at equimolar concentrations and RNA extended by the addition of ATP (B) or UTP (D).