Figure 1.
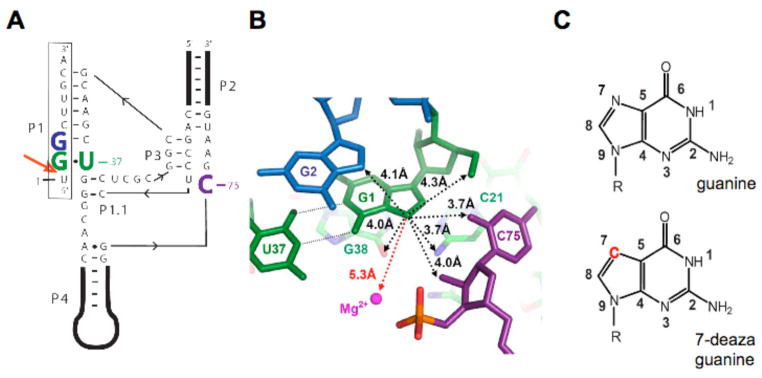
Overview of the genomic HDV ribozyme structure. (A) Secondary structure of the genomic HDV ribozyme used in this study. The 9-nt substrate strand (boxed) is introduced in trans. The solid line with embedded arrow shows the 5′ to 3′ direction of chain connectivity in the ribozyme strand. The cleavage site between U(−1) and G1 is indicated with a heavy red arrow. Important nucleotides are colored as follows: C75 is purple, the cleavage site G•U wobble pair is green and G2 is blue. (B) Atoms surrounding the G1·U37 wobble pair in post-cleaved HDV ribozyme (PDB code1cx0)(5). Nucleotides are colored as in panel a. Non-hydrogen atoms within 4.5 Å to the N7 of G1 are shown by black dotted lines. The crystallographic metal ion found closest to G1 is 5.3 Å away and highlighted by a red dashed line. (c) The chemical structures of guanosine and 7-deazaguanosine. Modification in 7-deazaguanine relative to guanine is highlighted in red.