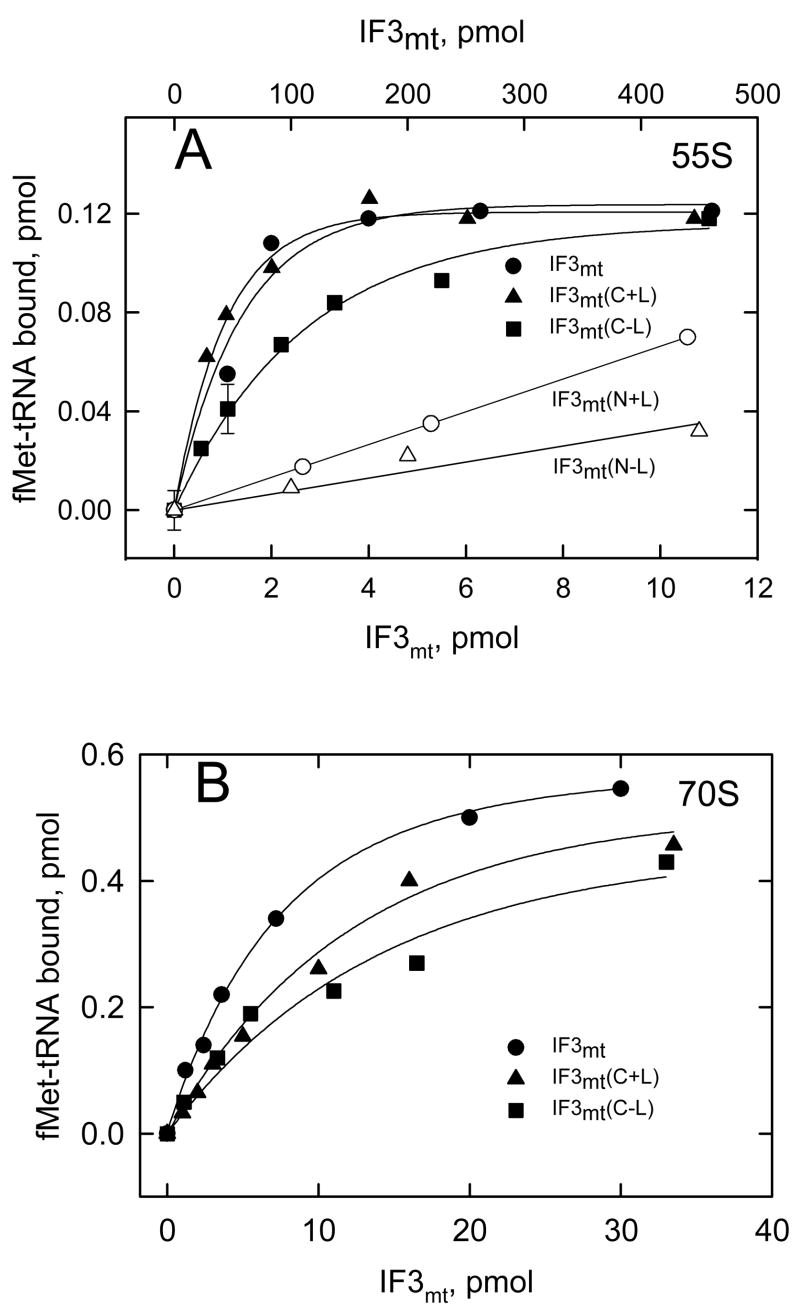
Figure 6. Effect of IF3mt and its C- and N-domain with and without the linker on initiation complex formation.
(A) [35S]fMet-tRNA binding to mitochondrial 55S particles (5 pmol, 50 nM) was tested in the presence of different concentrations of IF3mt and its C- and N-domain derivatives at a fixed amount of IF2mt (100 nM) using 10 μg poly(A,U,G) as the mRNA. A blank (about 0.18 pmol) representing the amount of label retained on the filter in the absence of IF3mt but in the presence of IF2mt has been subtracted from each value. The lower x-axis corresponds to full length IF3mt and its C-domain derivatives and the upper x-axis corresponds to the N-domain derivatives. (B) [35S]fMet-tRNA binding to E. coli 70S particles (20 pmol, 200 nM) was tested in the presence of different amounts of IF3mt and its C-domain derivatives at a fixed concentration of IF2mt (100 nM) using poly(A,U,G) as the mRNA. A blank representing the amount of label retained on the filter in the absence of IF3mt has been subtracted from each value (0.54 pmol). Representative errors are shown in panel A.