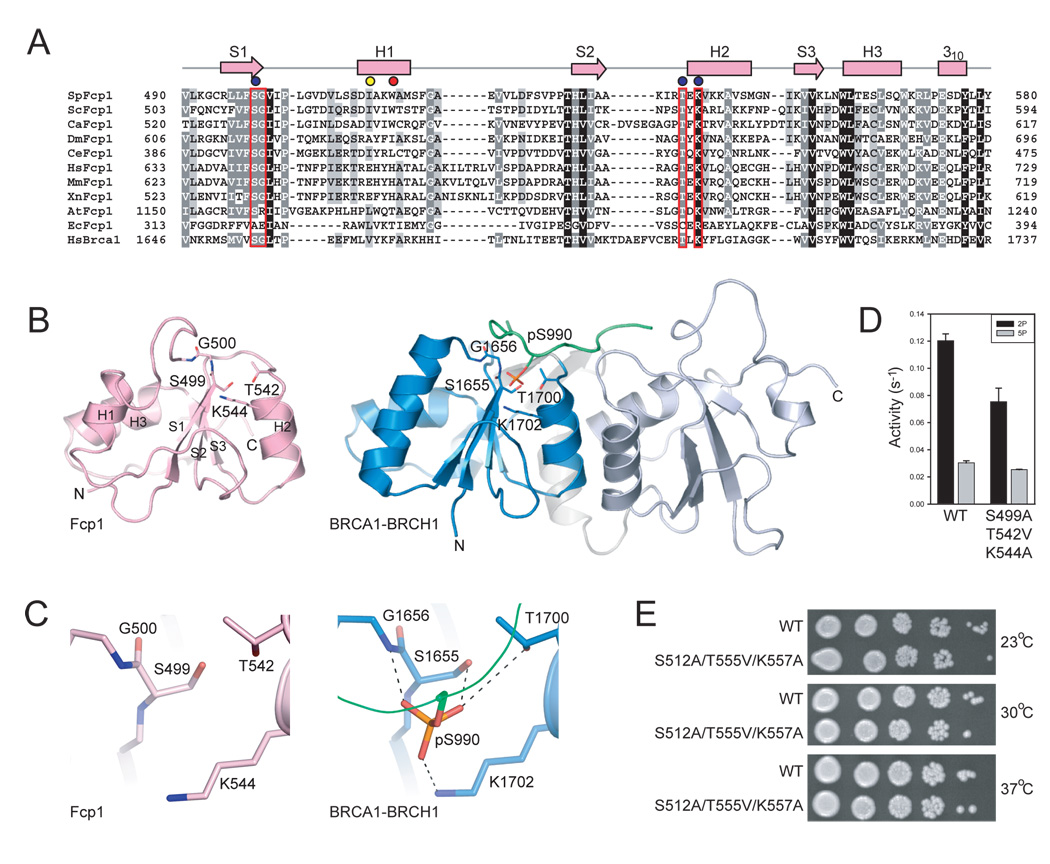
Figure 4. Fcp1 BRCT domain and comparison to BRCA1.
A) Structure-based amino acid alignment of the SpFcp1 BRCT domain to the proximal BRCT domain of human BRCA1 protein expanded to include BRCT domains of nine other Fcp1 orthologs with SpFcp1 secondary structure elements above the sequence. Gaps indicated by (−). Side chain identity/similarity denoted by shading (black conserved in all; grey conserved in most). BRCA1 amino acids that coordinate the phosphoserine residue of BRCH1 (panel B) outlined in red. Results of mutational analysis indicated by color-coded circles as in Fig. 2. B) Ribbon diagram for SpFcp1 BRCT (left) and human BRCA1 tandem BRCT domains bound to a BRCH1 phosphopeptide (green; phospho-serine stick representation; PDB 1T29). N and C indicate N- and C-terminal residues for BRCT domains, respectively. C) Close-up views of the putative phosphopeptide binding site in SpFcp1 and the actual phosphoserine binding site in BRCA1. Side chain contacts to phosphoserine shown as dashed lines. D) Phosphatase activities of wild-type Fcp1 and Fcp1-S499A/T542V/K544A with triheptad S2P and S5P CTD substrates. Error bars (one standard deviation) calculated from three independent experiments performed in triplicate. E) Serial dilutions of S. cerevisiae fcp1Δ strains bearing indicated FCP1 alleles spotted on YPAD agar grown at 23°C (top), 30°C (middle), and 37°C (bottom).