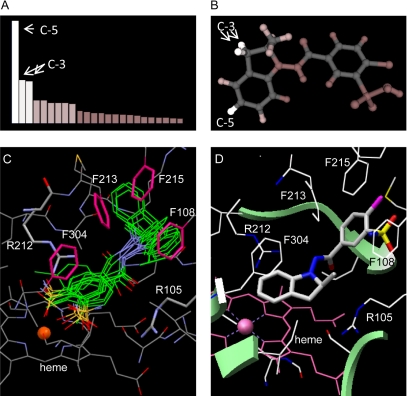
Fig. 7.
Predictions of indapamide metabolism by CYP3A4 with MetaSite (A and B), and molecular docking of indapamide in the active site of CYP3A4 by GOLD (C) and DOCK (D). The highest ranked hydrogen atoms (C-5 and C-3) by MetaSite that would most probably be metabolized are illustrated in A and B. Ten conformations of indapamide with the highest ranked fitness scores, as calculated by Goldscore function in GOLD, are illustrated by SILVER in overlapped green lines (C), which show the rotational trends of both the indoline ring and the chlorosulfamoylbenzamide moieties when docked in the active site of CYP3A4. The close distance between the two aromatic rings of indapamide and the phenylalanine cluster (Phe213, Phe215, and Phe108), as well as the Phe304 residue in the I-helix, indicate the domination of hydrophobic interactions. Phenylalanine rings are shown as pink sticks (C). The guanidinium moiety of Arg212 and Arg105 (in light blue sticks) are within hydrogen bonding distance of the benzamide oxygen (C). The highest ranked conformation of indapamide by DOCK is illustrated in B by Chimera using the anchor-and-grow algorithm in DOCK, in which the anchor is rigidly oriented, spatially clustered, and prioritized by its scoring function before the remaining flexible portion of indapamide is built. The conformation shows that the C-5 position of the indapamide indoline ring is the closest position to the heme iron (D). The heme is shown in pink with the iron as a sphere, and indapamide as color-coded sticks: nitrogen = blue, oxygen = red, chlorine = purple, and sulfur = yellow.