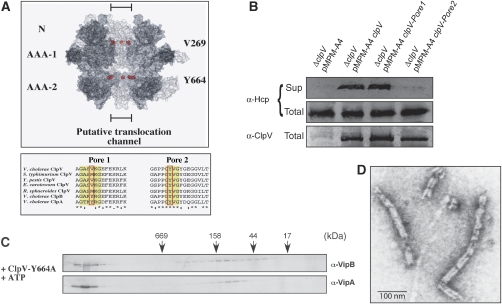
Figure 6.
A conserved pore-located residue of ClpV is crucial for T6S and remodelling of the VipA/VipB complex. (A) Upper part: surface representation of a ClpV hexamer model. Two monomers were removed from the hexamer to show the inside of the putative translocation channel. The positioning of N-terminal domains (N) and ATPase domains (AAA-1 and AAA-2) are indicated. Pore-located Val269 and Tyr664 are shown as CPK models and are coloured in red. Lower part: multiple sequence alignment of polypeptide segments located at the central pore entrance of the first or second AAA domain (pore 1 and pore 2, respectively) of various ClpV proteins. The protein sequence of the pore regions of V. cholerae ClpV was aligned with numerous other ClpV proteins from Salmonella typhimurium, Yersinia pestis, Erwinia carotowora and Rhodobacter sphaeroides. V. cholerae ClpA and ClpB are included as a reference. Conserved, pore-located residues that have been implicated in ClpA- or ClpB-mediated substrate threading are highlighted (red rectangle). (B) Complementation analysis of the V.c. ΔclpV secretion defect by plasmid-encoded clpV and pore mutant derivatives using Hcp-specific antibodies. ClpV levels were monitored by immunoblot analysis using ClpV-specific antibodies. Sup: culture supernatant, total: total cell extract. The ClpV-Y664A pore mutant does not efficiently dissociate VipA/VipB complexes as analysed by size-exclusion chromatography (C) and electron microscopy (D). A scale bar is indicated.