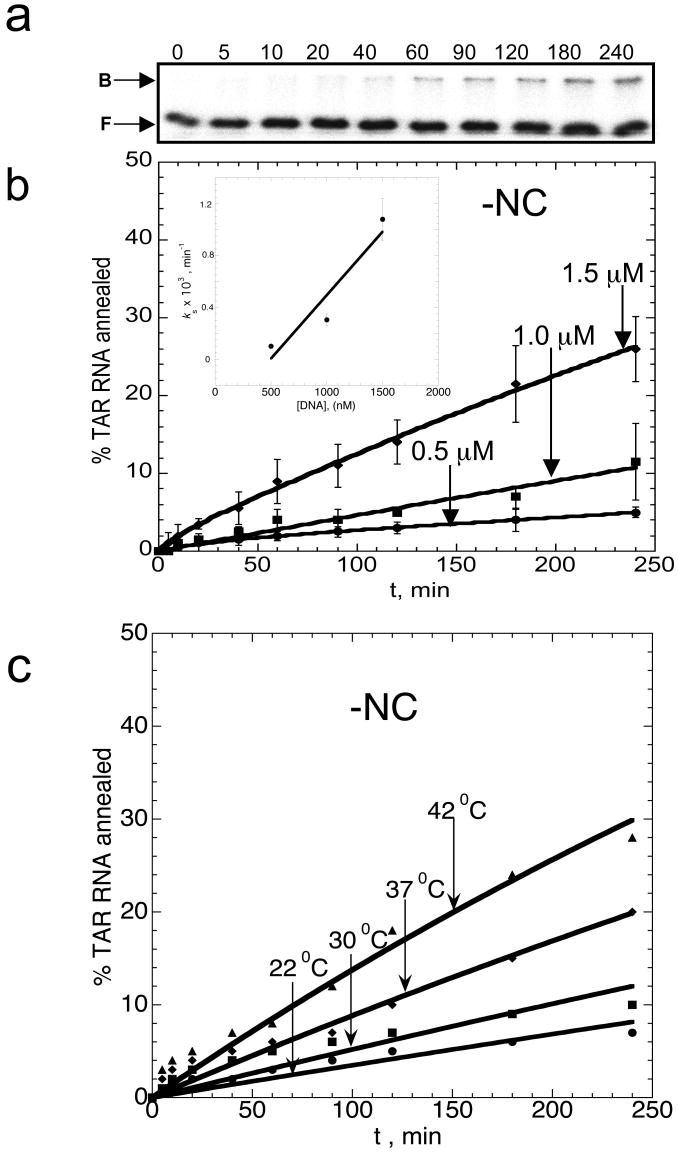
Figure 2.
Time course of TAR RNA/DNA hairpin annealing in the absence of NC. (a) Typical gel-shift analysis performed with 50 nM TAR RNA and 1 μM TAR DNA as a function of time. The numbers at the top of each lane correspond to minutes. Bound (B) and free (F) RNA bands are labeled on the left. (b) Time course showing the percent of TAR RNA (15 nM) annealed to variable concentrations of TAR DNA (as indicated on each curve) at 37 °C. Lines represent double-exponential fits of the data to equation (2), with the equilibrium percent annealed fixed at 100%. The insert shows the dominant slow annealing rate, ks obtained from the fits of the data shown in (b), as a function of TAR DNA concentration, D. The line is a linear fit of the data with a slope corresponding to keff=10±5 M-1s-1. (c) Temperature dependence of TAR RNA/DNA annealing. Time courses for TAR RNA (15 nM) annealing to TAR DNA (1.5 μM) were measured in the absence of NC in 20 mM Na+ at the indicated temperatures.