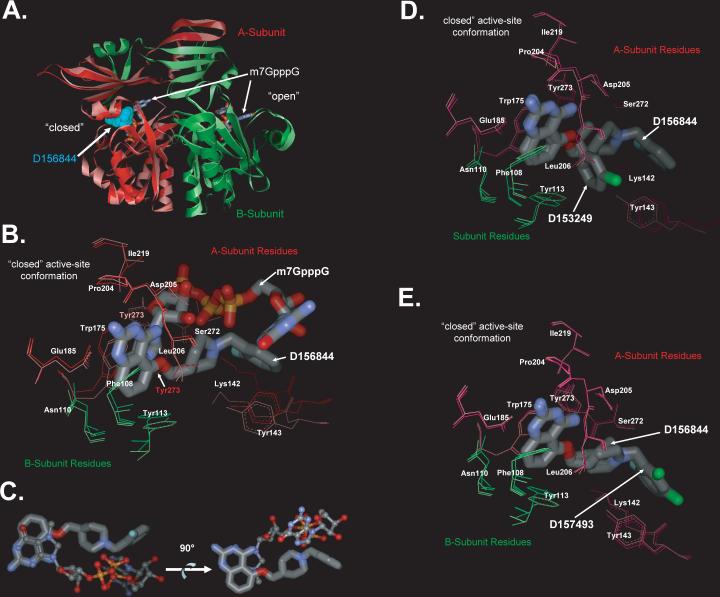
Figure 6. Crystal Structures of DcpS Bound to C5-quinazolines.
(A) Superposition of DcpS with bound D156844 and m7GpppG. The structure of DcpS bound to D156844 (cyan CPK space filling model) to human DcpS (pink Subunit A, and light green Subunit B) has been superimposed onto the previously reported structure of human DcpS (dark red Subunit A, and dark green Subunit B) with m7GpppG (stick model) bound to both the closed (left side) and open (right side) active site regions (PDB ID:1ST0) 29. The superposition illustrates that D156844 is bound only in closed active site conformation state, while m7GpppG is capable of binding into both the open and closed active site pockets. The DcpS structures with D156844 and m7GpppG bound superimpose with and RMSD on all atoms of 1.1 Å indicating that the conformations of the two are very similar.
(B) Superposition of D156844 and m7GpppG in the DcpS active site. The structure of DcpS bound to D156844 (labeled stick model) to human DcpS (pink Subunit A, and light green Subunit B) has been superimposed onto the previously reported structure of human DcpS (dark red Subunit A, and dark green Subunit B) with m7GpppG (labeled stick model) bound active site regions (PDB ID:1ST0) 29. The “closed” active-site conformation of DcpS is shown. Amino acid side chains are numbered and the ligands are identified. The 2−4 diamino quinazoline of for D156844 ligand adopts an identical binding mode as the m7G group of m7GpppG in DcpS. The C5-substituent of D156844 takes on a different conformation in binding to DcpS as compared to the pppG portion of the m7GpppG. Side chains which take on significantly different conformations between the two structures are Tyr273 and Lys142.
(C) Comparison of the binding pose of D156844 to m7GpppG cap substrate. The DcpS protein structures of our C5-quinazoline bound structure and that of the m7GpppG bound structure (PDB ID:1ST0) 12 were superimposed with COOT 29. The superposition the DcpS protein structures revealed that the quinazoline moiety D156844 (thick ball and stick) overlays nicely with that of the m7G moiety of m7GpppG (thin ball and stick) of the 1ST0 structure. In contrast, the C5 substituent of the quinazoline follows a different trajectory in the binding pocket compared to the pppG appendage of m7GpppG. The two images are rotated 90 degrees with respect to each other and the protein has been removed.
(D) Superposition of DcpS with bound D156844 and D153249. The “closed” active-site conformation of DcpS is shown in superposition mode for the DcpS co-crystal structures with D156844 (light pink for A subunit residues and light green for B subunit residues) and D153249 (dark pink for A subunit residues and dark green for B subunit residues). Amino acid side chains are numbered and the ligands are identified. The 2−4 diamino quinazoline of for each ligand adopts an identical binding mode in DcpS. The C5-substituent of each ligand takes on a different conformation in binding to DcpS.
(E) Superposition of DcpS with bound D156844 and D157493. The “closed” active-site conformation of DcpS is shown in superposition mode for the DcpS co-crystal structures with D156844 (light pink for A subunit residues and light green for B subunit residues) and D157493 (dark pink for A subunit residues and dark green for B subunit residues). Amino acid side chains are numbered and the ligands are identified. The 2−4 diamino quinazoline of for each ligand adopts an identical binding mode in DcpS. The C5-substituent of each ligand takes on a different conformation in binding to DcpS.