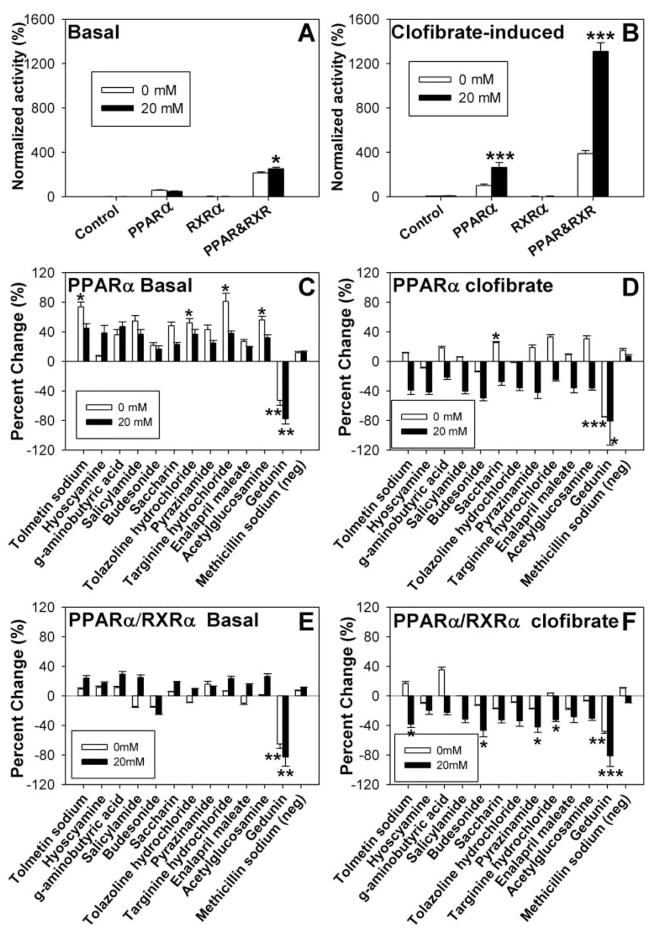
FIG. 2.
Glucose and compound effects on peroxisome proliferator-activated receptor-α (PPARα) transactivation. COS-7 cells transfected with PPARα, retinoid X receptor-α (RXRα), PPARα and RXRα, or pSG5 empty vector were analyzed for basal (A) and clofibrate-induced (B) transactivation of an acyl-CoA oxidase reporter construct in the absence (open bars) and presence of 20 mM glucose (filled bars). Transactivation values are presented as the percentage firefly luciferase activity normalized to Renilla luciferase (internal control), where clofibrate-induced PPARα activity in the absence of ligand is arbitrarily set to 100%. COS-7 cells transfected with PPARα (C, D) or PPARα and RXRα ( E, F ) were examined for compound effect on basal (C, E) and clofibrate-induced (D, F) transactivation of the acyl-CoA oxidase reporter construct in the absence (open bars) and presence (filled bars) of 20 mM glucose. Transactivation values are presented as the mean percentage change from the no-compound control ± SEM (n = 3). Asterisks (*) represent significant differences as compared with the no-glucose control for panels A and B or the no-compound control for panels C through F. *p < 0.05, **p < 0.01, ***p < 0.001.