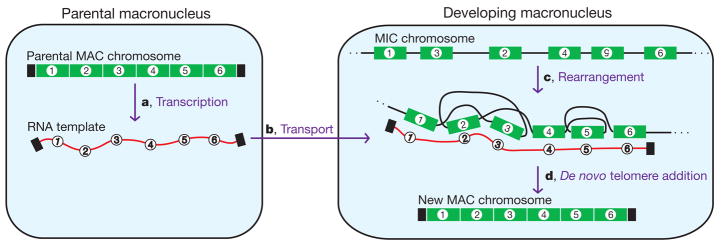
Figure 5. Model for RNA guiding of genome rearrangements during macronuclear development in Oxytricha.
a, Bidirectional RNA transcription of all DNA nanochromosomes (including injected DNA) in the old, maternal macronucleus (MAC) before its degradation. b, Transport of these RNA transcripts to the newly developing macronucleus, where they may act as scaffolds to guide rearrangements (deletion, permutation and inversion) of corresponding micronuclear (MIC) DNA sequences (c). This step would be notable and unprecedented, but perhaps possible if there were either local or extensive strand-separation of both the RNA template and the developing DNA (see ref. 20). In this illustration, segments 2 and 3 are switched and segment 5 is inverted (number upside down). d, De novo telomere addition (black rectangles) and amplification completes formation of new macronuclear nanochromosomes.