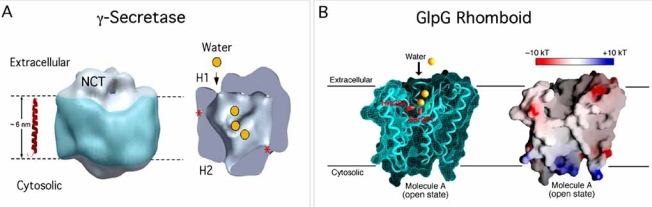
Fig. (4).
Three-dimensional structure of γ-secretase and GlpG, an E.coli intramembrane protease of the Rhomboid family. (A) Electron microscopic three-dimensional representation of the γ-secretase complex at a resolution of 15 Å. Left: The potential transmembrane segment with the belt-like structure (blue) is outlined by two parallel dashed lines, 60 Å apart, representing the membrane bilayer. The NCT ectodomain is located at the top density region as confirmed by lectin binding experiments. Right: A cut-open view of the complex from the side reveals a large central chamber of 20-40 Å in length, one opening (H1, 20 Å) facing the extracellular region and one opening facing the cytosolic region (H2, 20 Å). Two weak density regions are labeled with stars. Figure modified from [22] with permission (copyright National Academy of Sciences, USA). (B) Crystal structure of GlpG a bacterial rhomboid, in an open conformation and at a resolution of 2.6 Å. The putative catalytic-dyad Ser 201 and His 254 residues are shown in red. The water molecules (orange spheres) required to accomplish peptide bond hydrolysis may enter the membrane-embedded active sites by different routes from the outside (and/or inside?) aqueous solution, including the central chamber (GlpG) and the H1 and/or H2 pores (γ-secretase). Figure modified from [27] with permission.