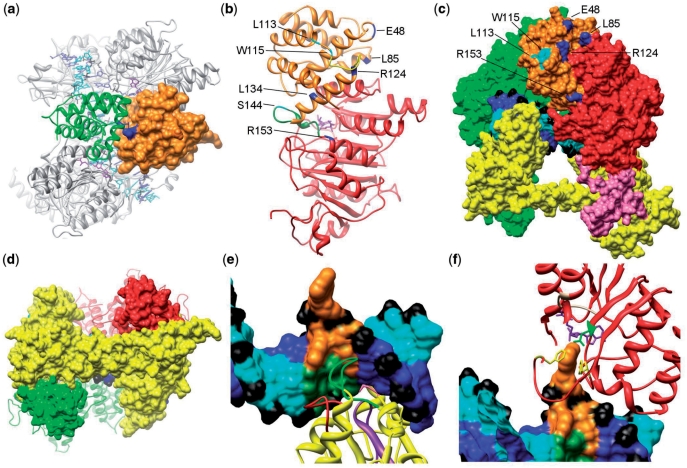
Figure 3.
(a) A top view of the M.EcoKI–DNA model highlighting the interface between the N-terminal domains of HsdM subunits. One N-terminal domain is shown as an orange surface and the second as a green ribbon. The R72Q mutation giving an r+m* phenotype is shown in blue and lies on the HsdM interface. (b) The m* mutations in the HsdM subunit. The N-terminal domain of HsdM is coloured orange and the central methyltransferase catalytic domain in red (the C-terminal domain is omitted). Residues where mutations cause the r−m* and r+m* phenotypes are shown coloured cyan and blue, respectively. Two flexible loops, disordered in the crystal structure, are shown in green and yellow. Both are associated with several m* residues. The cofactor AdoMet, in purple, can be seen in the catalytic domain. (c) The m* mutations in N-terminal domain of HsdM subunit presented in the M.EcoKI–DNA model. The HsdS subunit is yellow, the rear HsdM subunit as a green surface. The domains of the HsdM in the foreground are coloured as in Figure 2. The labelled residues in this HsdM subunit that cause r−m* and r+m* phenotypes are coloured cyan and blue, respectively. DNA is coloured blue and cyan with phosphates in black. (d) The contact between coiled-coil region of HsdS subunit and the C-terminal domain of HsdM subunit in M.EcoKI–DNA model. HsdS subunit surface is shown in yellow with the coiled-coil running horizontally across the image, HsdM subunits are coloured red and green with their C-terminal domains shown as a surface and the remainder as a ribbon. The close contact between the ends of the coiled-coil in HsdS with the C-terminal domains of HsdM can be seen. (e) Recognition of DNA by the N-terminal TRD of HsdS. Non-specific DNA is coloured blue and cyan with phosphates in black. The sequence AAC with the flipped out second A is shown in orange and the complementary sequence in dark green. The HsdS subunit is shown as a yellow ribbon with the previously identified DNA-contacting loops aa 83–91 in red and aa 96–118 in purple. These loops contact the complementary strand. The previously identified contact by Y27 (pink) can just be seen pointing into the minor groove at the rear of the image. The newly identified loop S139AGANINNIK148 is in green and primarily contacts the AAC sequence. (f) Contacts between the catalytic site of HsdM and DNA. DNA is shown as in (e) with the flipped out base sandwiched between HsdM F269 in the ‘NPPF’ motif IV and F345, both shown in yellow. The AdoMet cofactor is shown in purple and the HsdM motif I (beige) containing G177 is contacting the AdoMet. The rest of HsdM is in red. Two phenylalanine residues (F269 and F345, yellow) are available to form stacking interactions with the flipped out DNA base, and N266 (green) is within range for hydrogen bonding.