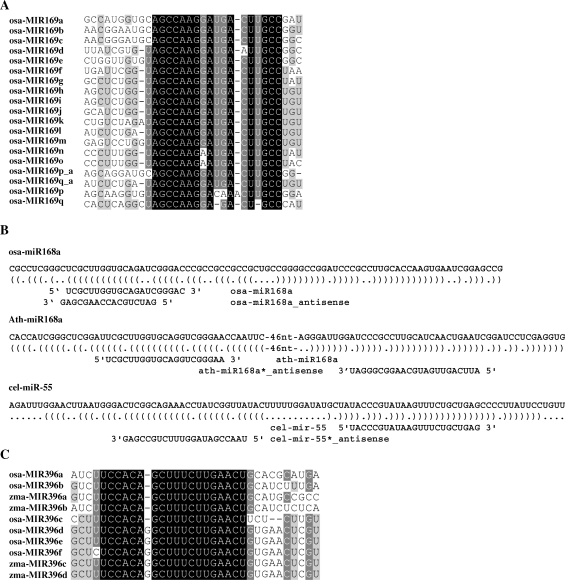
Figure 2.
Small RNAs match miRNA precursors. (A) The directions of osa-MIR169p and osa-MIR169q may be mis-annotated. Annotated precursor sequences of osa-miR169 family, together with reverse-complemented sequences of osa-MIR169p and osa-MIR169q, were aligned. osa-MIR169p_a and osa-MIR169q_a indicated the reverse complemented sequences of osa-MIR169p and osa-MIR169q. ClustalW and GeneDoc were used for sequences alignment and visualizing the conserved fragments. Black, dark gray and light gray shading indicated that the nucleotides were conserved in all, more than 80% (<100%), or more than 60% (<80%), of the sequences, respectively (same for C). (C) sRNAs matched mature miRNA or miRNA* in antisense orientation. The miRNA hairpins were shown in bracket notation. Precursors and mature miRNAs were shown in 5′ to 3′ orientation, whereas antisense sRNAs were shown in 3′ to 5′orientation. A. Subgroups of miR396 family in rice and maize. osa-miR396f and zma-miR396c/d were identified in this study.