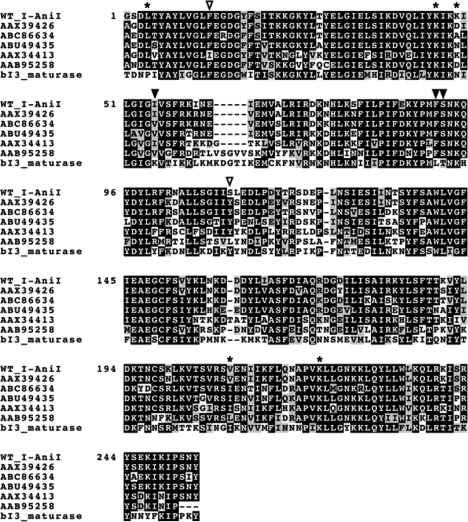
Figure 2.
Sequence alignment of I-AniI homologs. The alignment was performed using CLUSTALW program. Open triangles indicate positions of F13Y and S111Y in the Y2 I-AniI construct. AAX39426, ABC86634, ABU49435, AAX34413, AAB95258 and bI3 maturase are LAGLIDADG proteins from Neosartorya fischeri, Gibberella zeae, Phaeosphaeria nodorum SN15, Agrocybe chaxingu, Venturia inaequalis and Saccharomyces cerevisiae, respectively. Closed triangles represent sites of additional mutations I55V, F91I and S92T in the M5 I-AniI construct. Asterisks indicate remaining positions of point mutations isolated in the cleavage activity screen as listed in Table 1.